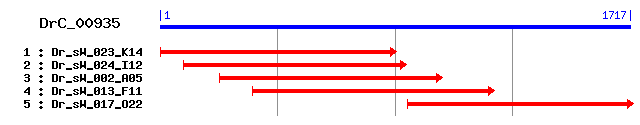
DrC_00935
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00935 (1717 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q920M5|CORO6_MOUSE Coronin-6 (Clipin E) 220 1e-56 sp|Q920J3|CORO6_RAT Coronin-6 216 1e-55 sp|Q9BR76|CO1B_HUMAN Coronin-1B (Coronin-2) 215 3e-55 sp|Q9WUM3|COR1B_MOUSE Coronin-1B (Coronin-2) 213 1e-54 sp|Q9ULV4|COR1C_HUMAN Coronin-1C (Coronin-3) (hCRNN4) 213 1e-54 sp|Q9WUM4|COR1C_MOUSE Coronin-1C (Coronin-3) 213 2e-54 sp|O89046|COR1B_RAT Coronin-1B (Coronin-2) 212 3e-54 sp|Q9XS70|COR1B_RABIT Coronin-1B (Coroninse) (Coronin-like ... 211 5e-54 sp|O89053|COR1A_MOUSE Coronin-1A (Coronin-like protein p57)... 200 1e-50 sp|Q92176|COR1A_BOVIN Coronin-1A (Coronin-like protein p57)... 199 3e-50
>sp|Q920M5|CORO6_MOUSE Coronin-6 (Clipin E) Length = 471 Score = 220 bits (560), Expect = 1e-56 Identities = 127/370 (34%), Positives = 202/370 (54%), Gaps = 2/370 (0%) Frame = +2 Query: 113 AVNAKYLAVLINCSTVDVMI-RPLEEVGRVESDPKGIKAAISLPTCIEWCPRDDDLLLIG 289 AVN K+LA+++ I PL + GRV+ + + I+WCP +D+++ Sbjct: 41 AVNPKFLAIIVEAGGGGAFIVLPLAKTGRVDKNYPLVTGHTGPVLDIDWCPHNDNVIASA 100 Query: 290 QQNGEINLIRIPDGGVTEVINTDDSEIKWNCESGSILNAVWHPTVGNAVLTSCGSKFAGA 469 + + + +IPD T V N + I S + WHPT N +L++ G Sbjct: 101 SDDTTVMVWQIPD--YTPVRNITEPVITLEGHSKRVGILSWHPTARNVLLSAGG------ 152 Query: 470 GSSADTHLILWDTVKTEKLLEIK-LNNCYLTSLSFNYNGSKVVCTDNSHITRIIDLRSSE 646 D +I+W+ E LL + ++ + S+ +N NGS + T RIID R S+ Sbjct: 153 ----DNVIIIWNVGTGEVLLSLDDIHPDVIHSVCWNSNGSLLATTCKDKTLRIIDPRKSQ 208 Query: 647 PVLQEAMLHKKARRQNTVYLKNGLIFSTGYSSTTIREYALWDENDLSNPIELIDLDANSG 826 V + A H+ AR V+ +G + STG+S + R+ ALWD N+ P+ L ++D ++G Sbjct: 209 VVAERARPHEGARPLRAVFTADGKLLSTGFSRMSERQLALWDPNNFEEPVALQEMDTSNG 268 Query: 827 VLYPIYDADINVIYVWGKGDSIMSFFEVVYEPKPTIQYLCCYQSAKPQNGVIAMPKLGVN 1006 VL P YD D +++Y+ GKGDS + +FE+ EP P + YL + S +PQ G+ MPK G++ Sbjct: 269 VLLPFYDPDSSIVYLCGKGDSSIRYFEITEEP-PFVHYLNTFSSKEPQRGMGFMPKRGLD 327 Query: 1007 VAANELTRLFKFYANGFCEVIPIICPRREQGFSEEVYPPTPSRVPSLTIDEWINGADADP 1186 V+ E+ R +K + CE I + PR+ F +++YP TP P+L DEW++G DA+P Sbjct: 328 VSKCEIARFYKLHERK-CEPIIMTVPRKSDLFQDDLYPDTPGPEPALEADEWLSGQDAEP 386 Query: 1187 ILISLETGEV 1216 +LISL+ G V Sbjct: 387 VLISLKEGYV 396
>sp|Q920J3|CORO6_RAT Coronin-6 Length = 472 Score = 216 bits (551), Expect = 1e-55 Identities = 126/370 (34%), Positives = 200/370 (54%), Gaps = 2/370 (0%) Frame = +2 Query: 113 AVNAKYLAVLINCSTVDVMI-RPLEEVGRVESDPKGIKAAISLPTCIEWCPRDDDLLLIG 289 AVN K+LA+++ I PL + GRV+ + I I+WCP +D+++ Sbjct: 41 AVNPKFLAIIVEAGGGGAFIVLPLAKTGRVDKNYPLITGHTGPVLDIDWCPHNDNVIASP 100 Query: 290 QQNGEINLIRIPDGGVTEVINTDDSEIKWNCESGSILNAVWHPTVGNAVLTSCGSKFAGA 469 + + + +IPD T V N + I S + WHPT N +L++ G Sbjct: 101 SDDTTVMVWQIPD--YTPVRNITEPVITLEGHSKRVGILSWHPTARNVLLSAGG------ 152 Query: 470 GSSADTHLILWDTVKTEKLLEIK-LNNCYLTSLSFNYNGSKVVCTDNSHITRIIDLRSSE 646 D +I+W+ E LL + ++ + S+ +N NGS + T RIID R S+ Sbjct: 153 ----DNVIIIWNVGTGEVLLSLDDIHPDVIHSVCWNSNGSLLATTCKDKTLRIIDPRKSQ 208 Query: 647 PVLQEAMLHKKARRQNTVYLKNGLIFSTGYSSTTIREYALWDENDLSNPIELIDLDANSG 826 V + H+ R V+ + G IF+TG++ + RE LWD N+ P+ L ++D ++G Sbjct: 209 VVAERFAAHEGMRPMRAVFTRLGHIFTTGFTRMSQRELGLWDPNNFEEPVALQEMDTSNG 268 Query: 827 VLYPIYDADINVIYVWGKGDSIMSFFEVVYEPKPTIQYLCCYQSAKPQNGVIAMPKLGVN 1006 VL P YD D +++Y+ GKGDS + +FE+ EP P + YL + S +PQ G+ MPK G++ Sbjct: 269 VLLPFYDPDSSIVYLCGKGDSSIRYFEITDEP-PFVHYLNTFSSKEPQRGMGFMPKRGLD 327 Query: 1007 VAANELTRLFKFYANGFCEVIPIICPRREQGFSEEVYPPTPSRVPSLTIDEWINGADADP 1186 V+ E+ R +K + CE I + PR+ F +++YP TP P+L DEW++G DA+P Sbjct: 328 VSKCEIARFYKLHERK-CEPIVMTVPRKSDLFQDDLYPDTPGPEPALEADEWLSGQDAEP 386 Query: 1187 ILISLETGEV 1216 +LISL+ G V Sbjct: 387 VLISLKEGYV 396
>sp|Q9BR76|CO1B_HUMAN Coronin-1B (Coronin-2) Length = 489 Score = 215 bits (548), Expect = 3e-55 Identities = 123/383 (32%), Positives = 207/383 (54%), Gaps = 2/383 (0%) Frame = +2 Query: 113 AVNAKYLAVLINCSTVDV-MIRPLEEVGRVESDPKGIKAAISLPTCIEWCPRDDDLLLIG 289 AVN K+LAV++ S ++ PL + GR++ + I+WCP +D+++ G Sbjct: 42 AVNPKFLAVIVEASGGGAFLVLPLSKTGRIDKAYPTVCGHTGPVLDIDWCPHNDEVIASG 101 Query: 290 QQNGEINLIRIPDGGVTEVINTDDSEIKWNCESGSILNAVWHPTVGNAVLTSCGSKFAGA 469 ++ + + +IP+ G+T + ++ + + I+ WHPT N +L++ Sbjct: 102 SEDCTVMVWQIPENGLTSPLTEPVVVLEGHTKRVGII--AWHPTARNVLLSA-------- 151 Query: 470 GSSADTHLILWDTVKTEKLLEI-KLNNCYLTSLSFNYNGSKVVCTDNSHITRIIDLRSSE 646 D +++W+ E+L + L+ + ++S+N+NGS RIID R Sbjct: 152 --GCDNVVLIWNVGTAEELYRLDSLHPDLIYNVSWNHNGSLFCSACKDKSVRIIDPRRGT 209 Query: 647 PVLQEAMLHKKARRQNTVYLKNGLIFSTGYSSTTIREYALWDENDLSNPIELIDLDANSG 826 V + H+ AR ++L +G +F+TG+S + R+ ALWD +L P+ L +LD+++G Sbjct: 210 LVAEREKAHEGARPMRAIFLADGKVFTTGFSRMSERQLALWDPENLEEPMALQELDSSNG 269 Query: 827 VLYPIYDADINVIYVWGKGDSIMSFFEVVYEPKPTIQYLCCYQSAKPQNGVIAMPKLGVN 1006 L P YD D +V+YV GKGDS + +FE+ EP P I +L + S +PQ G+ +MPK G+ Sbjct: 270 ALLPFYDPDTSVVYVCGKGDSSIRYFEITEEP-PYIHFLNTFTSKEPQRGMGSMPKRGLE 328 Query: 1007 VAANELTRLFKFYANGFCEVIPIICPRREQGFSEEVYPPTPSRVPSLTIDEWINGADADP 1186 V+ E+ R +K + CE I + PR+ F +++YP T +L +EW++G DADP Sbjct: 329 VSKCEIARFYKLHERK-CEPIVMTVPRKSDLFQDDLYPDTAGPEAALEAEEWVSGRDADP 387 Query: 1187 ILISLETGEVSMESKSTTSNVQR 1255 ILISL E + SK + R Sbjct: 388 ILISLR--EAYVPSKQRDLKISR 408
Score = 34.7 bits (78), Expect = 0.88
Identities = 12/33 (36%), Positives = 23/33 (69%)
Frame = +1
Query: 7 IRHSKFKNIYGDPKKKEQCFDQIKLKVKGADAT 105
+R SKF++++G P K +QC++ I++ D+T
Sbjct: 7 VRQSKFRHVFGQPVKNDQCYEDIRVSRVTWDST 39
>sp|Q9WUM3|COR1B_MOUSE Coronin-1B (Coronin-2) Length = 484 Score = 213 bits (543), Expect = 1e-54 Identities = 123/383 (32%), Positives = 207/383 (54%), Gaps = 2/383 (0%) Frame = +2 Query: 113 AVNAKYLAVLINCSTVDV-MIRPLEEVGRVESDPKGIKAAISLPTCIEWCPRDDDLLLIG 289 AVN K+LAV++ S M+ PL + GR++ + I+WCP +D+++ G Sbjct: 42 AVNPKFLAVIVEASGGGAFMVLPLNKTGRIDKAYPTVCGHTGPVLDIDWCPHNDEVIASG 101 Query: 290 QQNGEINLIRIPDGGVTEVINTDDSEIKWNCESGSILNAVWHPTVGNAVLTSCGSKFAGA 469 ++ + + +IP+ G+T + ++ + + I+ WHPT N +L++ Sbjct: 102 SEDCTVMVWQIPENGLTSPLTEPVVVLEGHTKRVGIIT--WHPTARNVLLSA-------- 151 Query: 470 GSSADTHLILWDTVKTEKLLEI-KLNNCYLTSLSFNYNGSKVVCTDNSHITRIIDLRSSE 646 D +++W+ E+L + L+ + ++S+N+NGS RIID R Sbjct: 152 --GCDNVVLIWNVGTAEELYRLDSLHPDLIYNVSWNHNGSLFCSACKDKSVRIIDPRRGT 209 Query: 647 PVLQEAMLHKKARRQNTVYLKNGLIFSTGYSSTTIREYALWDENDLSNPIELIDLDANSG 826 V + H+ AR ++L +G +F+TG+S + R+ ALWD +L P+ L +LD+++G Sbjct: 210 LVAEREKAHEGARPMRAIFLADGKVFTTGFSRMSERQLALWDPENLEEPMALQELDSSNG 269 Query: 827 VLYPIYDADINVIYVWGKGDSIMSFFEVVYEPKPTIQYLCCYQSAKPQNGVIAMPKLGVN 1006 L P YD D +V+YV GKGDS + +FE+ EP P I +L + S +PQ G+ +MPK G+ Sbjct: 270 ALLPFYDPDTSVVYVCGKGDSSIRYFEITDEP-PYIHFLNTFTSKEPQRGMGSMPKRGLE 328 Query: 1007 VAANELTRLFKFYANGFCEVIPIICPRREQGFSEEVYPPTPSRVPSLTIDEWINGADADP 1186 V+ E+ R +K + CE I + PR+ F +++YP T +L ++W++G DA+P Sbjct: 329 VSKCEIARFYKLHERK-CEPIVMTVPRKSDLFQDDLYPDTAGPEAALEAEDWVSGQDANP 387 Query: 1187 ILISLETGEVSMESKSTTSNVQR 1255 ILISL E + SK V R Sbjct: 388 ILISLR--EAYVPSKQRDLKVSR 408
Score = 34.7 bits (78), Expect = 0.88
Identities = 12/33 (36%), Positives = 23/33 (69%)
Frame = +1
Query: 7 IRHSKFKNIYGDPKKKEQCFDQIKLKVKGADAT 105
+R SKF++++G P K +QC++ I++ D+T
Sbjct: 7 VRQSKFRHVFGQPVKNDQCYEDIRVSRVTWDST 39
>sp|Q9ULV4|COR1C_HUMAN Coronin-1C (Coronin-3) (hCRNN4) Length = 474 Score = 213 bits (542), Expect = 1e-54 Identities = 127/433 (29%), Positives = 227/433 (52%), Gaps = 6/433 (1%) Frame = +2 Query: 113 AVNAKYLAVLINCSTVDV-MIRPLEEVGRVESDPKGIKAAISLPTCIEWCPRDDDLLLIG 289 AVN +++A++I S ++ PL + GR++ + I+WCP +D ++ G Sbjct: 40 AVNPRFVAIIIEASGGGAFLVLPLHKTGRIDKSYPTVCGHTGPVLDIDWCPHNDQVIASG 99 Query: 290 QQNGEINLIRIPDGGVTEVINTDDSEIKWNCESGSILNAVWHPTVGNAVLTSCGSKFAGA 469 ++ + + +IP+ G+T ++ + + S + WHPT N +L++ Sbjct: 100 SEDCTVMVWQIPENGLT--LSLTEPVVILEGHSKRVGIVAWHPTARNVLLSA-------- 149 Query: 470 GSSADTHLILWDTVKTEKLLEIK-LNNCYLTSLSFNYNGSKVVCTDNSHITRIIDLRSSE 646 D +I+W+ E L+ + +++ + ++S+N NGS + R+ID R E Sbjct: 150 --GCDNAIIIWNVGTGEALINLDDMHSDMIYNVSWNRNGSLICTASKDKKVRVIDPRKQE 207 Query: 647 PVLQEAMLHKKARRQNTVYLKNGLIFSTGYSSTTIREYALWDENDLSNPIELIDLDANSG 826 V ++ H+ AR ++L +G +F+TG+S + R+ ALW+ ++ PI L ++D ++G Sbjct: 208 IVAEKEKAHEGARPMRAIFLADGNVFTTGFSRMSERQLALWNPKNMQEPIALHEMDTSNG 267 Query: 827 VLYPIYDADINVIYVWGKGDSIMSFFEVVYEPKPTIQYLCCYQSAKPQNGVIAMPKLGVN 1006 VL P YD D ++IY+ GKGDS + +FE+ E P + YL + S +PQ G+ MPK G++ Sbjct: 268 VLLPFYDPDTSIIYLCGKGDSSIRYFEITDE-SPYVHYLNTFSSKEPQRGMGYMPKRGLD 326 Query: 1007 VAANELTRLFKFYANGFCEVIPIICPRREQGFSEEVYPPTPSRVPSLTIDEWINGADADP 1186 V E+ R FK + CE I + PR+ F +++YP T +L +EW G +ADP Sbjct: 327 VNKCEIARFFKLHERK-CEPIIMTVPRKSDLFQDDLYPDTAGPEAALEAEEWFEGKNADP 385 Query: 1187 ILISLETGEV---SMESKSTTSNVQRPK-NAQQKINEVMLLIHQFNSDENGNEHSLSDYQ 1354 ILISL+ G + + + K N+ K A +K + + + ++ NE L + Sbjct: 386 ILISLKHGYIPGKNRDLKVVKKNILDSKPTANKKCDLISIPKKTTDTASVQNEAKLDEIL 445 Query: 1355 TLIKNIQEAITGE 1393 IK+I++ I + Sbjct: 446 KEIKSIKDTICNQ 458
Score = 32.3 bits (72), Expect = 4.4
Identities = 10/25 (40%), Positives = 19/25 (76%)
Frame = +1
Query: 7 IRHSKFKNIYGDPKKKEQCFDQIKL 81
+R SKF++++G K +QC+D I++
Sbjct: 5 VRQSKFRHVFGQAVKNDQCYDDIRV 29
>sp|Q9WUM4|COR1C_MOUSE Coronin-1C (Coronin-3) Length = 474 Score = 213 bits (541), Expect = 2e-54 Identities = 128/433 (29%), Positives = 227/433 (52%), Gaps = 6/433 (1%) Frame = +2 Query: 113 AVNAKYLAVLINCSTVDV-MIRPLEEVGRVESDPKGIKAAISLPTCIEWCPRDDDLLLIG 289 AVN +++A++I S ++ PL + GR++ + I+WCP +D ++ G Sbjct: 40 AVNPRFVAIIIEASGGGAFLVLPLHKTGRIDKSYPTVCGHTGPVLDIDWCPHNDQVIASG 99 Query: 290 QQNGEINLIRIPDGGVTEVINTDDSEIKWNCESGSILNAVWHPTVGNAVLTSCGSKFAGA 469 ++ + + +IP+ G+T ++ + + S + WHPT N +L++ Sbjct: 100 SEDCTVMVWQIPENGLT--LSLTEPVVILEGHSKRVGIVAWHPTARNVLLSA-------- 149 Query: 470 GSSADTHLILWDTVKTEKLLEIK-LNNCYLTSLSFNYNGSKVVCTDNSHITRIIDLRSSE 646 D +I+W+ E L+ + +++ + ++S++ NGS + R+ID R E Sbjct: 150 --GCDNAIIIWNVGTGEALINLDDMHSDMIYNVSWSRNGSLICTASKDKKVRVIDPRKQE 207 Query: 647 PVLQEAMLHKKARRQNTVYLKNGLIFSTGYSSTTIREYALWDENDLSNPIELIDLDANSG 826 V ++ H+ AR ++L +G +F+TG+S + R+ ALW+ ++ PI L ++D ++G Sbjct: 208 IVAEKEKAHEGARPMRAIFLADGNVFTTGFSRMSERQLALWNPKNMQEPIALHEMDTSNG 267 Query: 827 VLYPIYDADINVIYVWGKGDSIMSFFEVVYEPKPTIQYLCCYQSAKPQNGVIAMPKLGVN 1006 VL P YD D ++IY+ GKGDS + +FE+ E P + YL + S +PQ G+ MPK G++ Sbjct: 268 VLLPFYDPDTSIIYLCGKGDSSIRYFEITDE-SPYVHYLNTFSSKEPQRGMGYMPKRGLD 326 Query: 1007 VAANELTRLFKFYANGFCEVIPIICPRREQGFSEEVYPPTPSRVPSLTIDEWINGADADP 1186 V E+ R FK + CE I + PR+ F +++YP T +L +EW G +ADP Sbjct: 327 VNKCEIARFFKLHERK-CEPIIMTVPRKSDLFQDDLYPDTAGPEAALEAEEWFEGKNADP 385 Query: 1187 ILISLETGEV---SMESKSTTSNVQRPKNAQQKINEVMLLIHQ-FNSDENGNEHSLSDYQ 1354 ILISL+ G + + + K N+ K A K +E+ + ++ NE L + Sbjct: 386 ILISLKHGYIPGKNRDLKVVKKNILDSKPAANKKSELSCAPKKPTDTASVQNEAKLDEIL 445 Query: 1355 TLIKNIQEAITGE 1393 IK+I+E I + Sbjct: 446 KEIKSIKETICSQ 458
Score = 32.3 bits (72), Expect = 4.4
Identities = 10/25 (40%), Positives = 19/25 (76%)
Frame = +1
Query: 7 IRHSKFKNIYGDPKKKEQCFDQIKL 81
+R SKF++++G K +QC+D I++
Sbjct: 5 VRQSKFRHVFGQAVKNDQCYDDIRV 29
>sp|O89046|COR1B_RAT Coronin-1B (Coronin-2) Length = 484 Score = 212 bits (539), Expect = 3e-54 Identities = 125/385 (32%), Positives = 208/385 (54%), Gaps = 4/385 (1%) Frame = +2 Query: 113 AVNAKYLAVLINCSTVDV-MIRPLEEVGRVESDPKGIKAAISLPTCIEWCPRDDDLLLIG 289 AVN K+LAV++ S M+ PL + GR++ + I+WCP +D+++ G Sbjct: 42 AVNPKFLAVIVEASGGGAFMVLPLNKTGRIDKAYPTVCGHTGPVLDIDWCPHNDEVIASG 101 Query: 290 QQNGEINLIRIPDGGVTEVINTDDSEIKWNCESGSILNAVWHPTVGNAVLTSCGSKFAGA 469 ++ + + +IP+ G+T + ++ + + I+ WHPT N +L++ Sbjct: 102 SEDCTVMVWQIPENGLTSPLTEPVVVLEGHTKRVGIIT--WHPTARNVLLSA-------- 151 Query: 470 GSSADTHLILWDTVKTEKLLEI-KLNNCYLTSLSFNYNGSK--VVCTDNSHITRIIDLRS 640 D +++W+ E+L + L+ + ++S+N+NGS C D S RIID R Sbjct: 152 --GCDNVVLIWNVGTAEELYRLDSLHPDLIYNVSWNHNGSLFCTACKDKS--VRIIDPRR 207 Query: 641 SEPVLQEAMLHKKARRQNTVYLKNGLIFSTGYSSTTIREYALWDENDLSNPIELIDLDAN 820 V + H+ AR ++L +G +F+ G+S + R+ ALWD + P+ L +LD++ Sbjct: 208 GTLVAEREKAHEGARPMRAIFLADGKVFTAGFSRMSERQLALWDPENFEEPMALQELDSS 267 Query: 821 SGVLYPIYDADINVIYVWGKGDSIMSFFEVVYEPKPTIQYLCCYQSAKPQNGVIAMPKLG 1000 +G L P YD D +V+YV GKGDS + +FE+ EP P I +L + S +PQ G+ +MPK G Sbjct: 268 NGALLPFYDPDTSVVYVCGKGDSSIRYFEITDEP-PYIHFLNTFTSKEPQRGMGSMPKRG 326 Query: 1001 VNVAANELTRLFKFYANGFCEVIPIICPRREQGFSEEVYPPTPSRVPSLTIDEWINGADA 1180 + V+ E+ R +K + CE I + PR+ F +++YP T +L ++W++G DA Sbjct: 327 LEVSKCEIARFYKLHERK-CEPIVMTVPRKSDLFQDDLYPDTAGPDAALEAEDWVSGQDA 385 Query: 1181 DPILISLETGEVSMESKSTTSNVQR 1255 DPILISL E + SK V R Sbjct: 386 DPILISLR--EAYVPSKQRDLKVSR 408
Score = 34.7 bits (78), Expect = 0.88
Identities = 12/33 (36%), Positives = 23/33 (69%)
Frame = +1
Query: 7 IRHSKFKNIYGDPKKKEQCFDQIKLKVKGADAT 105
+R SKF++++G P K +QC++ I++ D+T
Sbjct: 7 VRQSKFRHVFGQPVKNDQCYEDIRVSRVTWDST 39
>sp|Q9XS70|COR1B_RABIT Coronin-1B (Coroninse) (Coronin-like protein pp66) Length = 486 Score = 211 bits (537), Expect = 5e-54 Identities = 124/383 (32%), Positives = 204/383 (53%), Gaps = 2/383 (0%) Frame = +2 Query: 113 AVNAKYLAVLINCSTVDV-MIRPLEEVGRVESDPKGIKAAISLPTCIEWCPRDDDLLLIG 289 AVN K+LAV++ S ++ PL + GR++ + IEWCP +D ++ G Sbjct: 42 AVNPKFLAVIVEASGGGAFLVLPLSKTGRIDKAYPTVCGHTGPVLDIEWCPHNDGVIASG 101 Query: 290 QQNGEINLIRIPDGGVTEVINTDDSEIKWNCESGSILNAVWHPTVGNAVLTSCGSKFAGA 469 ++ + + +IP+ G+T + ++ + + I+ WHPT N +L++ Sbjct: 102 SEDCTVMVWQIPEDGLTSPLTEPVVVLEGHTKRVGIVT--WHPTARNVLLSA-------- 151 Query: 470 GSSADTHLILWDTVKTEKLLEI-KLNNCYLTSLSFNYNGSKVVCTDNSHITRIIDLRSSE 646 D +++W+ E+L + L+ + ++S+N NGS RIID R Sbjct: 152 --GCDNVVLIWNVGTAEELYRLDSLHPDLIYNVSWNRNGSLFCSACKDKSVRIIDPRRGT 209 Query: 647 PVLQEAMLHKKARRQNTVYLKNGLIFSTGYSSTTIREYALWDENDLSNPIELIDLDANSG 826 V + H+ AR ++L +G +F+TG+S + R+ ALWD +L P+ L +LD+++G Sbjct: 210 LVAEREKAHEGARPMRAIFLADGKVFTTGFSRMSERQLALWDPENLEEPMALQELDSSNG 269 Query: 827 VLYPIYDADINVIYVWGKGDSIMSFFEVVYEPKPTIQYLCCYQSAKPQNGVIAMPKLGVN 1006 L P YD D +V+YV GKGDS + +FE+ EP P I +L + S +PQ GV +MPK G+ Sbjct: 270 ALLPFYDPDTSVVYVCGKGDSSIRYFEITDEP-PYIHFLNTFTSKEPQRGVGSMPKRGLE 328 Query: 1007 VAANELTRLFKFYANGFCEVIPIICPRREQGFSEEVYPPTPSRVPSLTIDEWINGADADP 1186 V+ E+ R +K + CE I + PR+ F +++YP T +L +EW++G DA P Sbjct: 329 VSKCEIARFYKLHERK-CEPIVMTVPRKSDLFQDDLYPDTAGPEAALEAEEWVSGRDAGP 387 Query: 1187 ILISLETGEVSMESKSTTSNVQR 1255 +LISL E + SK V R Sbjct: 388 VLISLR--EAYVPSKQRDLKVSR 408
Score = 34.7 bits (78), Expect = 0.88
Identities = 12/33 (36%), Positives = 23/33 (69%)
Frame = +1
Query: 7 IRHSKFKNIYGDPKKKEQCFDQIKLKVKGADAT 105
+R SKF++++G P K +QC++ I++ D+T
Sbjct: 7 VRQSKFRHVFGQPVKNDQCYEDIRVSRVTWDST 39
>sp|O89053|COR1A_MOUSE Coronin-1A (Coronin-like protein p57) (Coronin-like protein A) (CLIPINA) (Tryptophan aspartate-containing coat protein) (TACO) Length = 461 Score = 200 bits (508), Expect = 1e-50 Identities = 116/381 (30%), Positives = 201/381 (52%), Gaps = 3/381 (0%) Frame = +2 Query: 113 AVNAKYLAVLINCSTVDV-MIRPLEEVGRVESDPKGIKAAISLPTCIEWCPRDDDLLLIG 289 AVN K++A++ S ++ PL + GRV+ + + + I WCP +D+++ G Sbjct: 41 AVNPKFMALICEASGGGAFLVLPLGKTGRVDKNVPLVCGHTAPVLDIAWCPHNDNVIASG 100 Query: 290 QQNGEINLIRIPDGGVTEVINTDDSEIKWNCESGSILNAVWHPTVGNAVLTSCGSKFAGA 469 ++ + + IPDGG+ + ++ + + I+ WHPT N +L++ Sbjct: 101 SEDCTVMVWEIPDGGLVLPLREPVITLEGHTKRVGIV--AWHPTAQNVLLSA-------- 150 Query: 470 GSSADTHLILWDTVKTEKLLEI--KLNNCYLTSLSFNYNGSKVVCTDNSHITRIIDLRSS 643 D +++WD +L + ++ + S+ ++ +G+ + + R+I+ R Sbjct: 151 --GCDNVILVWDVGTGAAVLTLGPDVHPDTIYSVDWSRDGALICTSCRDKRVRVIEPRKG 208 Query: 644 EPVLQEAMLHKKARRQNTVYLKNGLIFSTGYSSTTIREYALWDENDLSNPIELIDLDANS 823 V ++ H+ R + V++ G I +TG+S + R+ ALWD L P+ L +LD +S Sbjct: 209 TVVAEKDRPHEGTRPVHAVFVSEGKILTTGFSRMSERQVALWDTKHLEEPLSLQELDTSS 268 Query: 824 GVLYPIYDADINVIYVWGKGDSIMSFFEVVYEPKPTIQYLCCYQSAKPQNGVIAMPKLGV 1003 GVL P +D D N++Y+ GKGDS + +FE+ E P + YL + S + Q G+ MPK G+ Sbjct: 269 GVLLPFFDPDTNIVYLCGKGDSSIRYFEITSE-APFLHYLSMFSSKESQRGMGYMPKRGL 327 Query: 1004 NVAANELTRLFKFYANGFCEVIPIICPRREQGFSEEVYPPTPSRVPSLTIDEWINGADAD 1183 V E+ R +K + CE I + PR+ F E++YPPT P+LT +EW+ G DA Sbjct: 328 EVNKCEIARFYKLHERK-CEPIAMTVPRKSDLFQEDLYPPTAGPDPALTAEEWLGGRDAG 386 Query: 1184 PILISLETGEVSMESKSTTSN 1246 P+LISL+ G V +S+ N Sbjct: 387 PLLISLKDGYVPPKSRELRVN 407
Score = 33.1 bits (74), Expect = 2.6
Identities = 9/25 (36%), Positives = 20/25 (80%)
Frame = +1
Query: 7 IRHSKFKNIYGDPKKKEQCFDQIKL 81
+R SKF++++G P K +QC++ +++
Sbjct: 6 VRSSKFRHVFGQPAKADQCYEDVRV 30
>sp|Q92176|COR1A_BOVIN Coronin-1A (Coronin-like protein p57) (Coronin-like protein A) (CLIPINA) (Tryptophan aspartate-containing coat protein) (TACO) Length = 461 Score = 199 bits (505), Expect = 3e-50 Identities = 117/381 (30%), Positives = 201/381 (52%), Gaps = 3/381 (0%) Frame = +2 Query: 113 AVNAKYLAVLINCSTVDV-MIRPLEEVGRVESDPKGIKAAISLPTCIEWCPRDDDLLLIG 289 AVN K++A++ S ++ PL + GRV+ + + + I WCP +D+++ G Sbjct: 41 AVNPKFVALICEASGGGAFLVLPLGKTGRVDKNVPMVCGHTAPVLDIAWCPHNDNVIASG 100 Query: 290 QQNGEINLIRIPDGGVTEVINTDDSEIKWNCESGSILNAVWHPTVGNAVLTSCGSKFAGA 469 ++ + + IPDGG+T + ++ + + I+ WHPT N +L++ Sbjct: 101 SEDCSVMVWEIPDGGLTLPLREPVVTLEGHTKRVGIV--AWHPTAQNVLLSA-------- 150 Query: 470 GSSADTHLILWDTVKTEKLLEI--KLNNCYLTSLSFNYNGSKVVCTDNSHITRIIDLRSS 643 D +++WD +L + ++ + S+ ++ +G+ + + RII+ R Sbjct: 151 --GCDNVILVWDVGTGVAVLTLGSDVHPDTIYSVDWSRDGALICTSCRDKRVRIIEPRKG 208 Query: 644 EPVLQEAMLHKKARRQNTVYLKNGLIFSTGYSSTTIREYALWDENDLSNPIELIDLDANS 823 V ++ H+ R V++ +G I +TG+S + R+ ALWD L P+ L +LD +S Sbjct: 209 TIVAEKDRPHEGTRPVRAVFVSDGKILTTGFSRMSERQVALWDTKHLEEPLSLQELDTSS 268 Query: 824 GVLYPIYDADINVIYVWGKGDSIMSFFEVVYEPKPTIQYLCCYQSAKPQNGVIAMPKLGV 1003 GVL P +D D N++Y+ GKGDS + +FE+ E P + YL + S + Q G+ MPK G+ Sbjct: 269 GVLLPFFDPDTNIVYLCGKGDSSIRYFEITSE-APFLHYLSMFSSKESQRGMGYMPKRGL 327 Query: 1004 NVAANELTRLFKFYANGFCEVIPIICPRREQGFSEEVYPPTPSRVPSLTIDEWINGADAD 1183 V E+ R +K + CE I + PR+ F E++YPPT +LT +EW+ G DA Sbjct: 328 EVNKCEIARFYKLHERK-CEPIAMTVPRKSDLFQEDLYPPTAGPDAALTAEEWLGGRDAG 386 Query: 1184 PILISLETGEVSMESKSTTSN 1246 P+LISL+ G V +S+ N Sbjct: 387 PLLISLKDGYVPPKSRELRVN 407
Score = 33.1 bits (74), Expect = 2.6
Identities = 9/25 (36%), Positives = 20/25 (80%)
Frame = +1
Query: 7 IRHSKFKNIYGDPKKKEQCFDQIKL 81
+R SKF++++G P K +QC++ +++
Sbjct: 6 VRSSKFRHVFGQPAKADQCYEDVRV 30
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 184,071,094
Number of Sequences: 369166
Number of extensions: 3868924
Number of successful extensions: 10805
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 10010
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10715
length of database: 68,354,980
effective HSP length: 116
effective length of database: 46,925,720
effective search space used: 21351202600
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail