DrC_00869
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
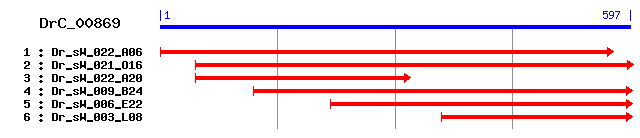
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00869 (597 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|O14880|MGST3_HUMAN Microsomal glutathione S-transferase ... 89 1e-17 sp|Q9CPU4|MGST3_MOUSE Microsomal glutathione S-transferase ... 89 1e-17 sp|P73795|Y1147_SYNY3 Hypothetical protein sll1147 54 3e-07 sp|Q16873|LTC4S_HUMAN Leukotriene C4 synthase (Leukotriene-... 37 0.038 sp|P55493|Y4IJ_RHISN Hypothetical 65.5 kDa protein y4iJ 32 1.6 sp|P27284|POLS_EEEV3 Structural polyprotein (P130) [Contain... 30 4.6 sp|O94477|MYO52_SCHPO Myosin-52 (Myosin type V-2) 30 6.0
>sp|O14880|MGST3_HUMAN Microsomal glutathione S-transferase 3 (Microsomal GST-3) (Microsomal GST-III) Length = 152 Score = 88.6 bits (218), Expect = 1e-17 Identities = 56/135 (41%), Positives = 76/135 (56%), Gaps = 3/135 (2%) Frame = +1 Query: 103 YGGVLMILAETYLLNFYLMYKVAKARKQYDVKCPTMYSDTSEE---FNCIQRSHQNYLEN 273 YG VL+ A ++++ +L V+KARK+Y V+ P MYS E FNCIQR+HQN LE Sbjct: 8 YGFVLLTGAASFIMVAHLAINVSKARKKYKVEYPIMYSTDPENGHIFNCIQRAHQNTLEV 67 Query: 274 ITFFIPFLLVSGLEFPRFTILCGKAFLIGRVLFDRGYTAEDPNKRKLGFKISLFGGLMPL 453 F+ FL V G+ PR G A+++GRVL+ GY +P+KR G G + L Sbjct: 68 YPPFLFFLAVGGVYHPRIASGLGLAWIVGRVLYAYGYYTGEPSKRSRGAL-----GSIAL 122 Query: 454 VGAVGVMAVGKCFLH 498 +G VG V F H Sbjct: 123 LGLVGT-TVCSAFQH 136
>sp|Q9CPU4|MGST3_MOUSE Microsomal glutathione S-transferase 3 (Microsomal GST-3) (Microsomal GST-III) Length = 153 Score = 88.6 bits (218), Expect = 1e-17 Identities = 55/122 (45%), Positives = 71/122 (58%), Gaps = 3/122 (2%) Frame = +1 Query: 103 YGGVLMILAETYLLNFYLMYKVAKARKQYDVKCPTMYSDTSEE---FNCIQRSHQNYLEN 273 YG VL+ A ++++ +L V KARK+Y V+ P MYS E FNCIQR+HQN LE Sbjct: 8 YGFVLLTGAASFVMVLHLAINVGKARKKYKVEYPVMYSTDPENGHMFNCIQRAHQNTLEV 67 Query: 274 ITFFIPFLLVSGLEFPRFTILCGKAFLIGRVLFDRGYTAEDPNKRKLGFKISLFGGLMPL 453 F+ FL V G+ PR G A++IGRVL+ GY DP+KR G SL L L Sbjct: 68 YPPFLFFLTVGGVYHPRIASGLGLAWIIGRVLYAYGYYTGDPSKRYRGAVGSL--ALFAL 125 Query: 454 VG 459 +G Sbjct: 126 MG 127
>sp|P73795|Y1147_SYNY3 Hypothetical protein sll1147 Length = 137 Score = 53.9 bits (128), Expect = 3e-07 Identities = 36/117 (30%), Positives = 60/117 (51%), Gaps = 1/117 (0%) Frame = +1 Query: 118 MILAETYLLNFYLMYKVAKARKQYDVKCPTMYSDTSEEFNCIQRSHQNYLENITFFIPFL 297 +I A +L L+ V +AR +Y V P + +E+F + R N LE + FF+P L Sbjct: 11 LITALATMLYLVLVINVGRARAKYGVMPPA--TTGNEDFERVLRVQYNTLEQLAFFLPGL 68 Query: 298 -LVSGLEFPRFTILCGKAFLIGRVLFDRGYTAEDPNKRKLGFKISLFGGLMPLVGAV 465 L + P + G +L+GR+L+ GY + KR +GF + ++ +VGA+ Sbjct: 69 WLFAIYRDPTIAAILGAVWLLGRILYAWGY-YQAAEKRMVGFALGSLSSMILVVGAL 124
>sp|Q16873|LTC4S_HUMAN Leukotriene C4 synthase (Leukotriene-C(4) synthase) (LTC4 synthase) Length = 150 Score = 37.0 bits (84), Expect = 0.038 Identities = 30/102 (29%), Positives = 47/102 (46%), Gaps = 1/102 (0%) Frame = +1 Query: 82 NEVAMLGYGGVLMILAETYLLNFYLMYKVAKARKQYDVKCPTMYSDTSEEFNCIQRSHQN 261 +EVA+L +L +L L Y +V AR+ + V P + EF + R+ N Sbjct: 3 DEVALLAAVTLLGVL-----LQAYFSLQVISARRAFRVSPPL--TTGPPEFERVYRAQVN 55 Query: 262 YLENITFFIPFLLVSGLEFPR-FTILCGKAFLIGRVLFDRGY 384 E F+ L V+G+ F LCG +L R+ + +GY Sbjct: 56 CSEYFPLFLATLWVAGIFFHEGAAALCGLVYLFARLRYFQGY 97
>sp|P55493|Y4IJ_RHISN Hypothetical 65.5 kDa protein y4iJ Length = 596 Score = 31.6 bits (70), Expect = 1.6 Identities = 16/49 (32%), Positives = 25/49 (51%) Frame = +1 Query: 274 ITFFIPFLLVSGLEFPRFTILCGKAFLIGRVLFDRGYTAEDPNKRKLGF 420 +TF +P+ ++ LE P FT+ G FL L G+ D + +GF Sbjct: 65 VTFSMPYEWLAALEQPTFTLTAGPPFLSSDYLDRFGFITADSSGLPVGF 113
>sp|P27284|POLS_EEEV3 Structural polyprotein (P130) [Contains: Coat protein C (Capsid protein C); Spike glycoprotein E3; Spike glycoprotein E2; 6 kDa peptide; Spike glycoprotein E1] Length = 1240 Score = 30.0 bits (66), Expect = 4.6 Identities = 22/76 (28%), Positives = 33/76 (43%), Gaps = 2/76 (2%) Frame = +1 Query: 202 PTMYSDTSEEFNCIQRSHQNYLENITFFIPFLLVSGLEFPRFTILCGKAFLI--GRVLFD 375 PT DT + N + ++QN+ T L+ + R CG AFL+ G Sbjct: 740 PTRADDTLQVLNYLWNNNQNFFWMQTLIPLAALIVCMRIVRCLFCCGPAFLLVCGAWAAA 799 Query: 376 RGYTAEDPNKRKLGFK 423 +TA PNK + +K Sbjct: 800 YEHTAVMPNKVGIPYK 815
>sp|O94477|MYO52_SCHPO Myosin-52 (Myosin type V-2) Length = 1516 Score = 29.6 bits (65), Expect = 6.0 Identities = 16/64 (25%), Positives = 32/64 (50%), Gaps = 3/64 (4%) Frame = +1 Query: 79 HNEVAMLGYGGVLMILAETY--LLNFYLMYKV-AKARKQYDVKCPTMYSDTSEEFNCIQR 249 +N++ + Y G+++I + L N Y V A + K D P +Y+ + + C+ + Sbjct: 97 YNQLQIYTYSGIVLIAVNPFQRLPNLYTHEIVRAYSEKSRDELDPHLYAIAEDSYKCMNQ 156 Query: 250 SHQN 261 H+N Sbjct: 157 EHKN 160
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 66,122,774
Number of Sequences: 369166
Number of extensions: 1282760
Number of successful extensions: 2943
Number of sequences better than 10.0: 7
Number of HSP's better than 10.0 without gapping: 2883
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2940
length of database: 68,354,980
effective HSP length: 105
effective length of database: 48,957,805
effective search space used: 4553075865
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail