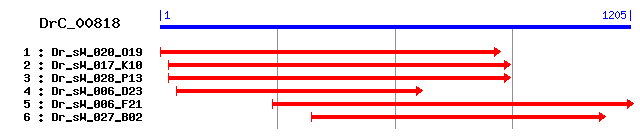
DrC_00818
- Cluster detail
- GO Category :
- biological_process :
- cellular_component :
- molecular_function :
- nucleotide binding
- ATP binding
- hydrogen-exporting ATPase activity, phosphorylative mechanism
- hydrogen ion transporter activity
- hydrolase activity
- nucleoside-triphosphatase activity
- metal ion binding
- hydrogen-transporting ATP synthase activity, rotational mechanism
- hydrogen-transporting ATPase activity, rotational mechanism
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00818 (1204 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P10719|ATPB_RAT ATP synthase beta chain, mitochondrial p... 445 e-124 sp|P00829|ATPB_BOVIN ATP synthase beta chain, mitochondrial... 445 e-124 sp|P56480|ATPB_MOUSE ATP synthase beta chain, mitochondrial... 444 e-124 sp|P06576|ATPB_HUMAN ATP synthase beta chain, mitochondrial... 442 e-124 sp|Q25117|ATPB_HEMPU ATP synthase beta chain, mitochondrial... 442 e-124 sp|Q9PTY0|ATPB_CYPCA ATP synthase beta chain, mitochondrial... 441 e-123 sp|P46561|ATPB_CAEEL ATP synthase beta chain, mitochondrial... 437 e-122 sp|Q05825|ATPB_DROME ATP synthase beta chain, mitochondrial... 429 e-119 sp|P00830|ATPB_YEAST ATP synthase beta chain, mitochondrial... 407 e-113 sp|P49376|ATPB_KLULA ATP synthase beta chain, mitochondrial... 401 e-111
>sp|P10719|ATPB_RAT ATP synthase beta chain, mitochondrial precursor Length = 529 Score = 445 bits (1145), Expect = e-124 Identities = 233/309 (75%), Positives = 257/309 (83%), Gaps = 4/309 (1%) Frame = +1 Query: 94 IRVLPNFVENKRNYATESVKKPEVSTKSNXXXXXXXXXXXXXXXHFDGDLPPILNALEVK 273 +R P V R+YA +S P+ T + FD LPPILNALEV+ Sbjct: 33 LRTAPAGVHPARDYAAQSSAAPKAGTATGQIVAVIGAVVDV---QFDEGLPPILNALEVQ 89 Query: 274 GRTPRLILEVAQHLGESTVRTIAMDGTEGLIRGQECLDVGSSIRIPVGPETLGRIMNVIG 453 GR RL+LEVAQHLGESTVRTIAMDGTEGL+RGQ+ LD G+ I+IPVGPETLGRIMNVIG Sbjct: 90 GRESRLVLEVAQHLGESTVRTIAMDGTEGLVRGQKVLDSGAPIKIPVGPETLGRIMNVIG 149 Query: 454 EPIDERGPIKSKMQSSIHQDAPDFVDMSTEQEILETGIKVVDLLAPYARGGKIGLFGGAG 633 EPIDERGPIK+K + IH +AP+F++MS EQEIL TGIKVVDLLAPYA+GGKIGLFGGAG Sbjct: 150 EPIDERGPIKTKQFAPIHAEAPEFIEMSVEQEILVTGIKVVDLLAPYAKGGKIGLFGGAG 209 Query: 634 VGKTVLIMELINNIARAHGGYSVFAGVGERTREGNDLYHEMITTGVIDLKGDKSKVSLVY 813 VGKTVLIMELINN+A+AHGGYSVFAGVGERTREGNDLYHEMI +GVI+LK SKV+LVY Sbjct: 210 VGKTVLIMELINNVAKAHGGYSVFAGVGERTREGNDLYHEMIESGVINLKDATSKVALVY 269 Query: 814 GQMNEPPGARARVALTGLTVAEYFRDQEGQDVLLFIDNIFRFTQAGSEVSALLGRIPSAV 993 GQMNEPPGARARVALTGLTVAEYFRDQEGQDVLLFIDNIFRFTQAGSEVSALLGRIPSAV Sbjct: 270 GQMNEPPGARARVALTGLTVAEYFRDQEGQDVLLFIDNIFRFTQAGSEVSALLGRIPSAV 329 Query: 994 G----LSTD 1008 G L+TD Sbjct: 330 GYQPTLATD 338
Score = 137 bits (345), Expect = 6e-32
Identities = 68/71 (95%), Positives = 69/71 (97%)
Frame = +2
Query: 992 LGYQPTLATDMGTMQERITTTNKGSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSRG 1171
+GYQPTLATDMGTMQERITTT KGSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSR
Sbjct: 329 VGYQPTLATDMGTMQERITTTKKGSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSRA 388
Query: 1172 IAELGIYPAVD 1204
IAELGIYPAVD
Sbjct: 389 IAELGIYPAVD 399
>sp|P00829|ATPB_BOVIN ATP synthase beta chain, mitochondrial precursor Length = 528 Score = 445 bits (1144), Expect = e-124 Identities = 232/309 (75%), Positives = 258/309 (83%), Gaps = 4/309 (1%) Frame = +1 Query: 94 IRVLPNFVENKRNYATESVKKPEVSTKSNXXXXXXXXXXXXXXXHFDGDLPPILNALEVK 273 +R P ++ R+YA ++ P+ + FD LPPILNALEV+ Sbjct: 33 LRAAPAALQPARDYAAQASPSPKAGATTGRIVAVIGAVVDV---QFDEGLPPILNALEVQ 89 Query: 274 GRTPRLILEVAQHLGESTVRTIAMDGTEGLIRGQECLDVGSSIRIPVGPETLGRIMNVIG 453 GR RL+LEVAQHLGESTVRTIAMDGTEGL+RGQ+ LD G+ IRIPVGPETLGRIMNVIG Sbjct: 90 GRETRLVLEVAQHLGESTVRTIAMDGTEGLVRGQKVLDSGAPIRIPVGPETLGRIMNVIG 149 Query: 454 EPIDERGPIKSKMQSSIHQDAPDFVDMSTEQEILETGIKVVDLLAPYARGGKIGLFGGAG 633 EPIDERGPIK+K ++IH +AP+FV+MS EQEIL TGIKVVDLLAPYA+GGKIGLFGGAG Sbjct: 150 EPIDERGPIKTKQFAAIHAEAPEFVEMSVEQEILVTGIKVVDLLAPYAKGGKIGLFGGAG 209 Query: 634 VGKTVLIMELINNIARAHGGYSVFAGVGERTREGNDLYHEMITTGVIDLKGDKSKVSLVY 813 VGKTVLIMELINN+A+AHGGYSVFAGVGERTREGNDLYHEMI +GVI+LK SKV+LVY Sbjct: 210 VGKTVLIMELINNVAKAHGGYSVFAGVGERTREGNDLYHEMIESGVINLKDATSKVALVY 269 Query: 814 GQMNEPPGARARVALTGLTVAEYFRDQEGQDVLLFIDNIFRFTQAGSEVSALLGRIPSAV 993 GQMNEPPGARARVALTGLTVAEYFRDQEGQDVLLFIDNIFRFTQAGSEVSALLGRIPSAV Sbjct: 270 GQMNEPPGARARVALTGLTVAEYFRDQEGQDVLLFIDNIFRFTQAGSEVSALLGRIPSAV 329 Query: 994 G----LSTD 1008 G L+TD Sbjct: 330 GYQPTLATD 338
Score = 137 bits (345), Expect = 6e-32
Identities = 68/71 (95%), Positives = 69/71 (97%)
Frame = +2
Query: 992 LGYQPTLATDMGTMQERITTTNKGSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSRG 1171
+GYQPTLATDMGTMQERITTT KGSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSR
Sbjct: 329 VGYQPTLATDMGTMQERITTTKKGSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSRA 388
Query: 1172 IAELGIYPAVD 1204
IAELGIYPAVD
Sbjct: 389 IAELGIYPAVD 399
>sp|P56480|ATPB_MOUSE ATP synthase beta chain, mitochondrial precursor Length = 529 Score = 444 bits (1142), Expect = e-124 Identities = 232/309 (75%), Positives = 257/309 (83%), Gaps = 4/309 (1%) Frame = +1 Query: 94 IRVLPNFVENKRNYATESVKKPEVSTKSNXXXXXXXXXXXXXXXHFDGDLPPILNALEVK 273 +R P V R+YA ++ P+ T + FD LPPILNALEV+ Sbjct: 33 LRAAPAGVHPARDYAAQASAAPKAGTATGRIVAVIGAVVDV---QFDEGLPPILNALEVQ 89 Query: 274 GRTPRLILEVAQHLGESTVRTIAMDGTEGLIRGQECLDVGSSIRIPVGPETLGRIMNVIG 453 GR RL+LEVAQHLGESTVRTIAMDGTEGL+RGQ+ LD G+ I+IPVGPETLGRIMNVIG Sbjct: 90 GRDSRLVLEVAQHLGESTVRTIAMDGTEGLVRGQKVLDSGAPIKIPVGPETLGRIMNVIG 149 Query: 454 EPIDERGPIKSKMQSSIHQDAPDFVDMSTEQEILETGIKVVDLLAPYARGGKIGLFGGAG 633 EPIDERGPIK+K + IH +AP+F++MS EQEIL TGIKVVDLLAPYA+GGKIGLFGGAG Sbjct: 150 EPIDERGPIKTKQFAPIHAEAPEFIEMSVEQEILVTGIKVVDLLAPYAKGGKIGLFGGAG 209 Query: 634 VGKTVLIMELINNIARAHGGYSVFAGVGERTREGNDLYHEMITTGVIDLKGDKSKVSLVY 813 VGKTVLIMELINN+A+AHGGYSVFAGVGERTREGNDLYHEMI +GVI+LK SKV+LVY Sbjct: 210 VGKTVLIMELINNVAKAHGGYSVFAGVGERTREGNDLYHEMIESGVINLKDATSKVALVY 269 Query: 814 GQMNEPPGARARVALTGLTVAEYFRDQEGQDVLLFIDNIFRFTQAGSEVSALLGRIPSAV 993 GQMNEPPGARARVALTGLTVAEYFRDQEGQDVLLFIDNIFRFTQAGSEVSALLGRIPSAV Sbjct: 270 GQMNEPPGARARVALTGLTVAEYFRDQEGQDVLLFIDNIFRFTQAGSEVSALLGRIPSAV 329 Query: 994 G----LSTD 1008 G L+TD Sbjct: 330 GYQPTLATD 338
Score = 137 bits (345), Expect = 6e-32
Identities = 68/71 (95%), Positives = 69/71 (97%)
Frame = +2
Query: 992 LGYQPTLATDMGTMQERITTTNKGSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSRG 1171
+GYQPTLATDMGTMQERITTT KGSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSR
Sbjct: 329 VGYQPTLATDMGTMQERITTTKKGSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSRA 388
Query: 1172 IAELGIYPAVD 1204
IAELGIYPAVD
Sbjct: 389 IAELGIYPAVD 399
>sp|P06576|ATPB_HUMAN ATP synthase beta chain, mitochondrial precursor Length = 529 Score = 442 bits (1138), Expect = e-124 Identities = 231/309 (74%), Positives = 256/309 (82%), Gaps = 4/309 (1%) Frame = +1 Query: 94 IRVLPNFVENKRNYATESVKKPEVSTKSNXXXXXXXXXXXXXXXHFDGDLPPILNALEVK 273 +R P V R+YA ++ P+ + FD LPPILNALEV+ Sbjct: 33 LRAAPTAVHPVRDYAAQTSPSPKAGAATGRIVAVIGAVVDV---QFDEGLPPILNALEVQ 89 Query: 274 GRTPRLILEVAQHLGESTVRTIAMDGTEGLIRGQECLDVGSSIRIPVGPETLGRIMNVIG 453 GR RL+LEVAQHLGESTVRTIAMDGTEGL+RGQ+ LD G+ I+IPVGPETLGRIMNVIG Sbjct: 90 GRETRLVLEVAQHLGESTVRTIAMDGTEGLVRGQKVLDSGAPIKIPVGPETLGRIMNVIG 149 Query: 454 EPIDERGPIKSKMQSSIHQDAPDFVDMSTEQEILETGIKVVDLLAPYARGGKIGLFGGAG 633 EPIDERGPIK+K + IH +AP+F++MS EQEIL TGIKVVDLLAPYA+GGKIGLFGGAG Sbjct: 150 EPIDERGPIKTKQFAPIHAEAPEFMEMSVEQEILVTGIKVVDLLAPYAKGGKIGLFGGAG 209 Query: 634 VGKTVLIMELINNIARAHGGYSVFAGVGERTREGNDLYHEMITTGVIDLKGDKSKVSLVY 813 VGKTVLIMELINN+A+AHGGYSVFAGVGERTREGNDLYHEMI +GVI+LK SKV+LVY Sbjct: 210 VGKTVLIMELINNVAKAHGGYSVFAGVGERTREGNDLYHEMIESGVINLKDATSKVALVY 269 Query: 814 GQMNEPPGARARVALTGLTVAEYFRDQEGQDVLLFIDNIFRFTQAGSEVSALLGRIPSAV 993 GQMNEPPGARARVALTGLTVAEYFRDQEGQDVLLFIDNIFRFTQAGSEVSALLGRIPSAV Sbjct: 270 GQMNEPPGARARVALTGLTVAEYFRDQEGQDVLLFIDNIFRFTQAGSEVSALLGRIPSAV 329 Query: 994 G----LSTD 1008 G L+TD Sbjct: 330 GYQPTLATD 338
Score = 137 bits (345), Expect = 6e-32
Identities = 68/71 (95%), Positives = 69/71 (97%)
Frame = +2
Query: 992 LGYQPTLATDMGTMQERITTTNKGSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSRG 1171
+GYQPTLATDMGTMQERITTT KGSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSR
Sbjct: 329 VGYQPTLATDMGTMQERITTTKKGSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSRA 388
Query: 1172 IAELGIYPAVD 1204
IAELGIYPAVD
Sbjct: 389 IAELGIYPAVD 399
>sp|Q25117|ATPB_HEMPU ATP synthase beta chain, mitochondrial precursor Length = 523 Score = 442 bits (1138), Expect = e-124 Identities = 225/264 (85%), Positives = 243/264 (92%), Gaps = 4/264 (1%) Frame = +1 Query: 229 FDGDLPPILNALEVKGRTPRLILEVAQHLGESTVRTIAMDGTEGLIRGQECLDVGSSIRI 408 F+ DLPPILNALEV+GRT RL+LEVAQHLGE+TVRTIAMDGTEGLIRGQ+C+D GS I I Sbjct: 70 FEDDLPPILNALEVQGRTSRLVLEVAQHLGENTVRTIAMDGTEGLIRGQKCVDTGSPISI 129 Query: 409 PVGPETLGRIMNVIGEPIDERGPIKSKMQSSIHQDAPDFVDMSTEQEILETGIKVVDLLA 588 PVGPETLGRI+NVIGEPIDERGPI + +S+IH +AP+FV+MS QEIL TGIKVVDLLA Sbjct: 130 PVGPETLGRIINVIGEPIDERGPIGTDRRSAIHAEAPEFVEMSVNQEILVTGIKVVDLLA 189 Query: 589 PYARGGKIGLFGGAGVGKTVLIMELINNIARAHGGYSVFAGVGERTREGNDLYHEMITTG 768 PYA+GGKIGLFGGAGVGKTVLIMELINN+A+AHGGYSVFAGVGERTREGNDLYHEMI G Sbjct: 190 PYAKGGKIGLFGGAGVGKTVLIMELINNVAKAHGGYSVFAGVGERTREGNDLYHEMIEGG 249 Query: 769 VIDLKGDKSKVSLVYGQMNEPPGARARVALTGLTVAEYFRDQEGQDVLLFIDNIFRFTQA 948 VI LK D SKV+LVYGQMNEPPGARARVALTGLTVAEYFRDQEGQDVLLFIDNIFRFTQA Sbjct: 250 VISLKDDTSKVALVYGQMNEPPGARARVALTGLTVAEYFRDQEGQDVLLFIDNIFRFTQA 309 Query: 949 GSEVSALLGRIPSAVG----LSTD 1008 GSEVSALLGRIPSAVG L+TD Sbjct: 310 GSEVSALLGRIPSAVGYQPTLATD 333
Score = 139 bits (351), Expect = 1e-32
Identities = 69/71 (97%), Positives = 70/71 (98%)
Frame = +2
Query: 992 LGYQPTLATDMGTMQERITTTNKGSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSRG 1171
+GYQPTLATDMGTMQERITTT KGSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSRG
Sbjct: 324 VGYQPTLATDMGTMQERITTTKKGSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSRG 383
Query: 1172 IAELGIYPAVD 1204
IAELGIYPAVD
Sbjct: 384 IAELGIYPAVD 394
>sp|Q9PTY0|ATPB_CYPCA ATP synthase beta chain, mitochondrial precursor Length = 518 Score = 441 bits (1135), Expect = e-123 Identities = 225/264 (85%), Positives = 241/264 (91%), Gaps = 4/264 (1%) Frame = +1 Query: 229 FDGDLPPILNALEVKGRTPRLILEVAQHLGESTVRTIAMDGTEGLIRGQECLDVGSSIRI 408 FD DLPPILNALEV GR RL+LEVAQHLGE+TVRTIAMDGTEGL+RGQ+ LD G+ IRI Sbjct: 65 FDEDLPPILNALEVAGRDTRLVLEVAQHLGENTVRTIAMDGTEGLVRGQKVLDTGAPIRI 124 Query: 409 PVGPETLGRIMNVIGEPIDERGPIKSKMQSSIHQDAPDFVDMSTEQEILETGIKVVDLLA 588 PVGPETLGRIMNVIGEPIDERGPI +K + IH +AP+F DMS EQEIL TGIKVVDLLA Sbjct: 125 PVGPETLGRIMNVIGEPIDERGPITTKQTAPIHAEAPEFTDMSVEQEILVTGIKVVDLLA 184 Query: 589 PYARGGKIGLFGGAGVGKTVLIMELINNIARAHGGYSVFAGVGERTREGNDLYHEMITTG 768 PYA+GGKIGLFGGAGVGKTVLIMELINN+A+AHGGYSVFAGVGERTREGNDLYHEMI +G Sbjct: 185 PYAKGGKIGLFGGAGVGKTVLIMELINNVAKAHGGYSVFAGVGERTREGNDLYHEMIESG 244 Query: 769 VIDLKGDKSKVSLVYGQMNEPPGARARVALTGLTVAEYFRDQEGQDVLLFIDNIFRFTQA 948 VI+LK SKV+LVYGQMNEPPGARARVALTGLTVAEYFRDQEGQDVLLFIDNIFRFTQA Sbjct: 245 VINLKDTTSKVALVYGQMNEPPGARARVALTGLTVAEYFRDQEGQDVLLFIDNIFRFTQA 304 Query: 949 GSEVSALLGRIPSAVG----LSTD 1008 GSEVSALLGRIPSAVG L+TD Sbjct: 305 GSEVSALLGRIPSAVGYQPTLATD 328
Score = 137 bits (345), Expect = 6e-32
Identities = 68/71 (95%), Positives = 69/71 (97%)
Frame = +2
Query: 992 LGYQPTLATDMGTMQERITTTNKGSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSRG 1171
+GYQPTLATDMGTMQERITTT KGSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSR
Sbjct: 319 VGYQPTLATDMGTMQERITTTKKGSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSRA 378
Query: 1172 IAELGIYPAVD 1204
IAELGIYPAVD
Sbjct: 379 IAELGIYPAVD 389
>sp|P46561|ATPB_CAEEL ATP synthase beta chain, mitochondrial precursor Length = 538 Score = 437 bits (1125), Expect = e-122 Identities = 232/317 (73%), Positives = 254/317 (80%), Gaps = 12/317 (3%) Frame = +1 Query: 94 IRVLPNFVENKRNY--------ATESVKKPEVSTKSNXXXXXXXXXXXXXXXHFDGDLPP 249 IR+ N VE+K+ A + K +T +N FD +LPP Sbjct: 31 IRLSSNNVESKKGIHTGVATQQAAAATKVSAKATAANASGRIVAVIGAVVDVQFDENLPP 90 Query: 250 ILNALEVKGRTPRLILEVAQHLGESTVRTIAMDGTEGLIRGQECLDVGSSIRIPVGPETL 429 ILN LEV GR+PRLILEV+QHLG++ VR IAMDGTEGL+RGQ D G I+IPVGPETL Sbjct: 91 ILNGLEVVGRSPRLILEVSQHLGDNVVRCIAMDGTEGLVRGQPVADTGDPIKIPVGPETL 150 Query: 430 GRIMNVIGEPIDERGPIKSKMQSSIHQDAPDFVDMSTEQEILETGIKVVDLLAPYARGGK 609 GRIMNVIGEPIDERGPI SK ++IH +AP+FV+MS EQEIL TGIKVVDLLAPYA+GGK Sbjct: 151 GRIMNVIGEPIDERGPIASKNFAAIHAEAPEFVEMSVEQEILVTGIKVVDLLAPYAKGGK 210 Query: 610 IGLFGGAGVGKTVLIMELINNIARAHGGYSVFAGVGERTREGNDLYHEMITTGVIDLKGD 789 IGLFGGAGVGKTVLIMELINN+A+AHGGYSVFAGVGERTREGNDLYHEMI GVIDLKG Sbjct: 211 IGLFGGAGVGKTVLIMELINNVAKAHGGYSVFAGVGERTREGNDLYHEMIEGGVIDLKGK 270 Query: 790 KSKVSLVYGQMNEPPGARARVALTGLTVAEYFRDQEGQDVLLFIDNIFRFTQAGSEVSAL 969 SKVSLVYGQMNEPPGARARV LTGLTVAEYFRDQEGQDVLLFIDNIFRFTQAGSEVSAL Sbjct: 271 NSKVSLVYGQMNEPPGARARVCLTGLTVAEYFRDQEGQDVLLFIDNIFRFTQAGSEVSAL 330 Query: 970 LGRIPSAVG----LSTD 1008 LGRIPSAVG L+TD Sbjct: 331 LGRIPSAVGYQPTLATD 347
Score = 135 bits (341), Expect = 2e-31
Identities = 67/71 (94%), Positives = 69/71 (97%)
Frame = +2
Query: 992 LGYQPTLATDMGTMQERITTTNKGSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSRG 1171
+GYQPTLATDMG+MQERITTT KGSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSRG
Sbjct: 338 VGYQPTLATDMGSMQERITTTKKGSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSRG 397
Query: 1172 IAELGIYPAVD 1204
IAEL IYPAVD
Sbjct: 398 IAELAIYPAVD 408
>sp|Q05825|ATPB_DROME ATP synthase beta chain, mitochondrial precursor Length = 505 Score = 429 bits (1102), Expect = e-119 Identities = 220/264 (83%), Positives = 238/264 (90%), Gaps = 4/264 (1%) Frame = +1 Query: 229 FDGDLPPILNALEVKGRTPRLILEVAQHLGESTVRTIAMDGTEGLIRGQECLDVGSSIRI 408 FD +LPPILNALEV R+PRL+LEVAQHLGE+TVRTIAMDGTEGL+RGQ+ LD G IRI Sbjct: 52 FDDNLPPILNALEVDNRSPRLVLEVAQHLGENTVRTIAMDGTEGLVRGQKVLDTGYPIRI 111 Query: 409 PVGPETLGRIMNVIGEPIDERGPIKSKMQSSIHQDAPDFVDMSTEQEILETGIKVVDLLA 588 PVG ETLGRI+NVIGEPIDERGPI + ++IH +AP+FV MS EQEIL TGIKVVDLLA Sbjct: 112 PVGAETLGRIINVIGEPIDERGPIDTDKTAAIHAEAPEFVQMSVEQEILVTGIKVVDLLA 171 Query: 589 PYARGGKIGLFGGAGVGKTVLIMELINNIARAHGGYSVFAGVGERTREGNDLYHEMITTG 768 PYA+GGKIGLFGGAGVGKTVLIMELINN+A+AHGGYSVFAGVGERTREGNDLY+EMI G Sbjct: 172 PYAKGGKIGLFGGAGVGKTVLIMELINNVAKAHGGYSVFAGVGERTREGNDLYNEMIEGG 231 Query: 769 VIDLKGDKSKVSLVYGQMNEPPGARARVALTGLTVAEYFRDQEGQDVLLFIDNIFRFTQA 948 VI LK SKV+LVYGQMNEPPGARARVALTGLTVAEYFRDQEGQDVLLFIDNIFRFTQA Sbjct: 232 VISLKDKTSKVALVYGQMNEPPGARARVALTGLTVAEYFRDQEGQDVLLFIDNIFRFTQA 291 Query: 949 GSEVSALLGRIPSAVG----LSTD 1008 GSEVSALLGRIPSAVG L+TD Sbjct: 292 GSEVSALLGRIPSAVGYQPTLATD 315
Score = 135 bits (341), Expect = 2e-31
Identities = 67/71 (94%), Positives = 69/71 (97%)
Frame = +2
Query: 992 LGYQPTLATDMGTMQERITTTNKGSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSRG 1171
+GYQPTLATDMG+MQERITTT KGSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSR
Sbjct: 306 VGYQPTLATDMGSMQERITTTKKGSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSRA 365
Query: 1172 IAELGIYPAVD 1204
IAELGIYPAVD
Sbjct: 366 IAELGIYPAVD 376
>sp|P00830|ATPB_YEAST ATP synthase beta chain, mitochondrial precursor Length = 511 Score = 407 bits (1047), Expect = e-113 Identities = 210/266 (78%), Positives = 234/266 (87%), Gaps = 5/266 (1%) Frame = +1 Query: 226 HFD-GDLPPILNALEVKGRTPRLILEVAQHLGESTVRTIAMDGTEGLIRGQECLDVGSSI 402 HF+ +LP ILNALE+K +L+LEVAQHLGE+TVRTIAMDGTEGL+RG++ LD G I Sbjct: 57 HFEQSELPAILNALEIKTPQGKLVLEVAQHLGENTVRTIAMDGTEGLVRGEKVLDTGGPI 116 Query: 403 RIPVGPETLGRIMNVIGEPIDERGPIKSKMQSSIHQDAPDFVDMSTEQEILETGIKVVDL 582 +PVG ETLGRI+NVIGEPIDERGPIKSK++ IH D P F + ST EILETGIKVVDL Sbjct: 117 SVPVGRETLGRIINVIGEPIDERGPIKSKLRKPIHADPPSFAEQSTSAEILETGIKVVDL 176 Query: 583 LAPYARGGKIGLFGGAGVGKTVLIMELINNIARAHGGYSVFAGVGERTREGNDLYHEMIT 762 LAPYARGGKIGLFGGAGVGKTV I ELINNIA+AHGG+SVFAGVGERTREGNDLY EM Sbjct: 177 LAPYARGGKIGLFGGAGVGKTVFIQELINNIAKAHGGFSVFAGVGERTREGNDLYREMKE 236 Query: 763 TGVIDLKGDKSKVSLVYGQMNEPPGARARVALTGLTVAEYFRDQEGQDVLLFIDNIFRFT 942 TGVI+L+G+ SKV+LV+GQMNEPPGARARVALTGLT+AEYFRD+EGQDVLLFIDNIFRFT Sbjct: 237 TGVINLEGE-SKVALVFGQMNEPPGARARVALTGLTIAEYFRDEEGQDVLLFIDNIFRFT 295 Query: 943 QAGSEVSALLGRIPSAVG----LSTD 1008 QAGSEVSALLGRIPSAVG L+TD Sbjct: 296 QAGSEVSALLGRIPSAVGYQPTLATD 321
Score = 127 bits (319), Expect = 6e-29
Identities = 59/71 (83%), Positives = 69/71 (97%)
Frame = +2
Query: 992 LGYQPTLATDMGTMQERITTTNKGSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSRG 1171
+GYQPTLATDMG +QERITTT KGS+TSVQA+YVPADDLTDP+P+T+FAHLDA++VLSRG
Sbjct: 312 VGYQPTLATDMGLLQERITTTKKGSVTSVQAVYVPADDLTDPSPSTSFAHLDASSVLSRG 371
Query: 1172 IAELGIYPAVD 1204
I+ELGIYPAVD
Sbjct: 372 ISELGIYPAVD 382
>sp|P49376|ATPB_KLULA ATP synthase beta chain, mitochondrial precursor Length = 505 Score = 401 bits (1031), Expect = e-111 Identities = 207/262 (79%), Positives = 228/262 (87%), Gaps = 4/262 (1%) Frame = +1 Query: 235 GDLPPILNALEVKGRTPRLILEVAQHLGESTVRTIAMDGTEGLIRGQECLDVGSSIRIPV 414 G LP ILNALE+ +L+LEVAQHLGE+TVRTIAMDGTEGL+RG+ D G+ I +PV Sbjct: 55 GQLPAILNALEIDTPEGKLVLEVAQHLGENTVRTIAMDGTEGLVRGENVSDTGAPISVPV 114 Query: 415 GPETLGRIMNVIGEPIDERGPIKSKMQSSIHQDAPDFVDMSTEQEILETGIKVVDLLAPY 594 G ETLGRI+NVIGEPIDERGPI SKM+ IH D P FV+ ST E+LETGIKVVDLLAPY Sbjct: 115 GRETLGRIINVIGEPIDERGPINSKMRKPIHADPPLFVEQSTAAEVLETGIKVVDLLAPY 174 Query: 595 ARGGKIGLFGGAGVGKTVLIMELINNIARAHGGYSVFAGVGERTREGNDLYHEMITTGVI 774 ARGGKIGLFGGAGVGKTV I ELINNIA+AHGG+SVF GVGERTREGNDLY EM TGVI Sbjct: 175 ARGGKIGLFGGAGVGKTVFIQELINNIAKAHGGFSVFTGVGERTREGNDLYREMKETGVI 234 Query: 775 DLKGDKSKVSLVYGQMNEPPGARARVALTGLTVAEYFRDQEGQDVLLFIDNIFRFTQAGS 954 +L+GD SKV+LV+GQMNEPPGARARVALTGLT+AEYFRD+EGQDVLLFIDNIFRFTQAGS Sbjct: 235 NLEGD-SKVALVFGQMNEPPGARARVALTGLTIAEYFRDEEGQDVLLFIDNIFRFTQAGS 293 Query: 955 EVSALLGRIPSAVG----LSTD 1008 EVSALLGRIPSAVG L+TD Sbjct: 294 EVSALLGRIPSAVGYQPTLATD 315
Score = 134 bits (337), Expect = 5e-31
Identities = 64/71 (90%), Positives = 69/71 (97%)
Frame = +2
Query: 992 LGYQPTLATDMGTMQERITTTNKGSITSVQAIYVPADDLTDPAPATTFAHLDATTVLSRG 1171
+GYQPTLATDMG +QERITTT KGS+TSVQA+YVPADDLTDPAPATTFAHLDATTVLSRG
Sbjct: 306 VGYQPTLATDMGLLQERITTTKKGSVTSVQAVYVPADDLTDPAPATTFAHLDATTVLSRG 365
Query: 1172 IAELGIYPAVD 1204
I+ELGIYPAVD
Sbjct: 366 ISELGIYPAVD 376
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 127,554,772
Number of Sequences: 369166
Number of extensions: 2517979
Number of successful extensions: 8088
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 7060
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7655
length of database: 68,354,980
effective HSP length: 113
effective length of database: 47,479,925
effective search space used: 13626738475
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail