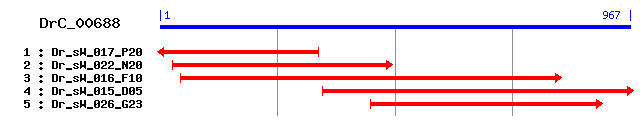
DrC_00688
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00688 (967 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P00940|TPIS_CHICK Triosephosphate isomerase (TIM) (Trios... 373 e-103 sp|P54714|TPIS_CANFA Triosephosphate isomerase (TIM) (Trios... 361 1e-99 sp|Q60HC9|TPIS_MACFA Triosephosphate isomerase (TIM) (Trios... 361 2e-99 sp|P60174|TPIS_HUMAN Triosephosphate isomerase (TIM) (Trios... 360 4e-99 sp|Q589R5|TPIS_ORYLA Triosephosphate isomerase (TIM) (Trios... 360 4e-99 sp|P00939|TPIS_RABIT Triosephosphate isomerase (TIM) (Trios... 358 1e-98 sp|P48500|TPIS_RAT Triosephosphate isomerase (TIM) (Triose-... 357 3e-98 sp|P17751|TPIS_MOUSE Triosephosphate isomerase (TIM) (Trios... 353 5e-97 sp|P48501|TPIS_SCHMA Triosephosphate isomerase (TIM) (Trios... 339 6e-93 sp|P00941|TPIS_LATCH Triosephosphate isomerase (TIM) (Trios... 337 3e-92
>sp|P00940|TPIS_CHICK Triosephosphate isomerase (TIM) (Triose-phosphate isomerase) Length = 248 Score = 373 bits (958), Expect = e-103 Identities = 182/244 (74%), Positives = 206/244 (84%) Frame = +3 Query: 57 RKFFVGGNWKMNGDKDELTKICNSLMNAKLHETTEVVVAPPSIYLDFVRQKLPAKIGVSA 236 RKFFVGGNWKMNGDK L ++ ++L AKL TEVV PSIYLDF RQKL AKIGV+A Sbjct: 4 RKFFVGGNWKMNGDKKSLGELIHTLNGAKLSADTEVVCGAPSIYLDFARQKLDAKIGVAA 63 Query: 237 QNCYKIPKGAFTGEISPAMIKDNMCDWVILGHSERRHVFGESDHLIGEKVKHALESGLSI 416 QNCYK+PKGAFTGEISPAMIKD WVILGHSERRHVFGESD LIG+KV HAL GL + Sbjct: 64 QNCYKVPKGAFTGEISPAMIKDIGAAWVILGHSERRHVFGESDELIGQKVAHALAEGLGV 123 Query: 417 IPCIGEKLDERESNKTNEVCFRQLKEIAANIKDWTRVVIAYEPVWAIGTGKTASPAQAQE 596 I CIGEKLDERE+ T +V F Q K IA N+KDW++VV+AYEPVWAIGTGKTA+P QAQE Sbjct: 124 IACIGEKLDEREAGITEKVVFEQTKAIADNVKDWSKVVLAYEPVWAIGTGKTATPQQAQE 183 Query: 597 VHKCVRDWLKTNVSEEVAKTTRILYGGSVTSGNCKELASQPDVDGFLVGGASLKPEFIDI 776 VH+ +R WLK++VS+ VA++TRI+YGGSVT GNCKELASQ DVDGFLVGGASLKPEF+DI Sbjct: 184 VHEKLRGWLKSHVSDAVAQSTRIIYGGSVTGGNCKELASQHDVDGFLVGGASLKPEFVDI 243 Query: 777 INAK 788 INAK Sbjct: 244 INAK 247
>sp|P54714|TPIS_CANFA Triosephosphate isomerase (TIM) (Triose-phosphate isomerase) Length = 249 Score = 361 bits (927), Expect = 1e-99 Identities = 175/248 (70%), Positives = 202/248 (81%) Frame = +3 Query: 45 MSNQRKFFVGGNWKMNGDKDELTKICNSLMNAKLHETTEVVVAPPSIYLDFVRQKLPAKI 224 M+ RKFFVGGNWKMNG K L ++ +L AK+ TEVV APP+ Y+DF RQKL AKI Sbjct: 1 MAPSRKFFVGGNWKMNGRKKNLGELITTLNAAKVPADTEVVCAPPTAYIDFARQKLDAKI 60 Query: 225 GVSAQNCYKIPKGAFTGEISPAMIKDNMCDWVILGHSERRHVFGESDHLIGEKVKHALES 404 V+AQNCYK+ GAFTGEISP MIKD WV+LGHSERRHVFGESD LIG+KV HAL Sbjct: 61 AVAAQNCYKVTNGAFTGEISPGMIKDCGATWVVLGHSERRHVFGESDELIGQKVAHALAE 120 Query: 405 GLSIIPCIGEKLDERESNKTNEVCFRQLKEIAANIKDWTRVVIAYEPVWAIGTGKTASPA 584 GL +I CIGEKLDERE+ T +V F Q K IA N+KDW++VV+AYEPVWAIGTGKTA+P Sbjct: 121 GLGVIACIGEKLDEREAGITEKVVFEQTKVIADNVKDWSKVVLAYEPVWAIGTGKTATPQ 180 Query: 585 QAQEVHKCVRDWLKTNVSEEVAKTTRILYGGSVTSGNCKELASQPDVDGFLVGGASLKPE 764 QAQEVH+ +R WLK+NVS+ VA++TRI+YGGSVT CKELASQPDVDGFLVGGASLKPE Sbjct: 181 QAQEVHEKLRGWLKSNVSDAVAQSTRIIYGGSVTGATCKELASQPDVDGFLVGGASLKPE 240 Query: 765 FIDIINAK 788 F+DIINAK Sbjct: 241 FVDIINAK 248
>sp|Q60HC9|TPIS_MACFA Triosephosphate isomerase (TIM) (Triose-phosphate isomerase) sp|P15426|TPIS_MACMU Triosephosphate isomerase (TIM) (Triose-phosphate isomerase) Length = 249 Score = 361 bits (926), Expect = 2e-99 Identities = 175/248 (70%), Positives = 201/248 (81%) Frame = +3 Query: 45 MSNQRKFFVGGNWKMNGDKDELTKICNSLMNAKLHETTEVVVAPPSIYLDFVRQKLPAKI 224 M+ RKFFVGGNWKMNG K L ++ +L AK+ TEVV APP+ Y+DF RQKL KI Sbjct: 1 MAPSRKFFVGGNWKMNGRKQNLGELIGTLNAAKVPADTEVVCAPPTAYIDFARQKLDPKI 60 Query: 225 GVSAQNCYKIPKGAFTGEISPAMIKDNMCDWVILGHSERRHVFGESDHLIGEKVKHALES 404 V+AQNCYK+ GAFTGEISP MIKD WV+LGHSERRHVFGESD LIG+KV HAL Sbjct: 61 AVAAQNCYKVTNGAFTGEISPGMIKDCGATWVVLGHSERRHVFGESDELIGQKVAHALAE 120 Query: 405 GLSIIPCIGEKLDERESNKTNEVCFRQLKEIAANIKDWTRVVIAYEPVWAIGTGKTASPA 584 GL +I CIGEKLDERE+ T +V F Q K IA N+KDW++VV+AYEPVWAIGTGKTA+P Sbjct: 121 GLGVIACIGEKLDEREAGITEKVVFEQTKVIADNVKDWSKVVLAYEPVWAIGTGKTATPQ 180 Query: 585 QAQEVHKCVRDWLKTNVSEEVAKTTRILYGGSVTSGNCKELASQPDVDGFLVGGASLKPE 764 QAQEVH+ +R WLK+NVSE VA++TRI+YGGSVT CKELASQPDVDGFLVGGASLKPE Sbjct: 181 QAQEVHEKLRGWLKSNVSEAVAQSTRIIYGGSVTGATCKELASQPDVDGFLVGGASLKPE 240 Query: 765 FIDIINAK 788 F+DIINAK Sbjct: 241 FVDIINAK 248
>sp|P60174|TPIS_HUMAN Triosephosphate isomerase (TIM) (Triose-phosphate isomerase) sp|P60175|TPIS_PANTR Triosephosphate isomerase (TIM) (Triose-phosphate isomerase) Length = 249 Score = 360 bits (923), Expect = 4e-99 Identities = 174/248 (70%), Positives = 201/248 (81%) Frame = +3 Query: 45 MSNQRKFFVGGNWKMNGDKDELTKICNSLMNAKLHETTEVVVAPPSIYLDFVRQKLPAKI 224 M+ RKFFVGGNWKMNG K L ++ +L AK+ TEVV APP+ Y+DF RQKL KI Sbjct: 1 MAPSRKFFVGGNWKMNGRKQSLGELIGTLNAAKVPADTEVVCAPPTAYIDFARQKLDPKI 60 Query: 225 GVSAQNCYKIPKGAFTGEISPAMIKDNMCDWVILGHSERRHVFGESDHLIGEKVKHALES 404 V+AQNCYK+ GAFTGEISP MIKD WV+LGHSERRHVFGESD LIG+KV HAL Sbjct: 61 AVAAQNCYKVTNGAFTGEISPGMIKDCGATWVVLGHSERRHVFGESDELIGQKVAHALAE 120 Query: 405 GLSIIPCIGEKLDERESNKTNEVCFRQLKEIAANIKDWTRVVIAYEPVWAIGTGKTASPA 584 GL +I CIGEKLDERE+ T +V F Q K IA N+KDW++VV+AYEPVWAIGTGKTA+P Sbjct: 121 GLGVIACIGEKLDEREAGITEKVVFEQTKVIADNVKDWSKVVLAYEPVWAIGTGKTATPQ 180 Query: 585 QAQEVHKCVRDWLKTNVSEEVAKTTRILYGGSVTSGNCKELASQPDVDGFLVGGASLKPE 764 QAQEVH+ +R WLK+NVS+ VA++TRI+YGGSVT CKELASQPDVDGFLVGGASLKPE Sbjct: 181 QAQEVHEKLRGWLKSNVSDAVAQSTRIIYGGSVTGATCKELASQPDVDGFLVGGASLKPE 240 Query: 765 FIDIINAK 788 F+DIINAK Sbjct: 241 FVDIINAK 248
>sp|Q589R5|TPIS_ORYLA Triosephosphate isomerase (TIM) (Triose-phosphate isomerase) Length = 248 Score = 360 bits (923), Expect = 4e-99 Identities = 172/245 (70%), Positives = 204/245 (83%) Frame = +3 Query: 54 QRKFFVGGNWKMNGDKDELTKICNSLMNAKLHETTEVVVAPPSIYLDFVRQKLPAKIGVS 233 QR+FFVGGNWKMNG+K+ L ++ ++L A LHE TEVV A PSIYLDF R L ++I V+ Sbjct: 3 QRRFFVGGNWKMNGNKESLEELMSTLNTASLHEDTEVVCAAPSIYLDFARSGLDSRISVA 62 Query: 234 AQNCYKIPKGAFTGEISPAMIKDNMCDWVILGHSERRHVFGESDHLIGEKVKHALESGLS 413 AQNCYK+ KGAFTGEISPAMIKD DWVILGHSERRHVFGE+D LIG+KV HALES LS Sbjct: 63 AQNCYKVAKGAFTGEISPAMIKDCGADWVILGHSERRHVFGENDELIGQKVAHALESDLS 122 Query: 414 IIPCIGEKLDERESNKTNEVCFRQLKEIAANIKDWTRVVIAYEPVWAIGTGKTASPAQAQ 593 +I CIGEKL+ERE+ T +V F Q + IA N+ DW +VV+AYEPVWAIGTGKTA+P QAQ Sbjct: 123 VIACIGEKLEEREAGTTEDVIFAQTQVIAENVMDWEKVVLAYEPVWAIGTGKTATPEQAQ 182 Query: 594 EVHKCVRDWLKTNVSEEVAKTTRILYGGSVTSGNCKELASQPDVDGFLVGGASLKPEFID 773 EVH+ +R W + N+SEEV+ + RI+YGGSVT+ NC+ELASQ DVDGFLVGGASLKPEFID Sbjct: 183 EVHEKLRAWFRANISEEVSDSLRIIYGGSVTAANCRELASQTDVDGFLVGGASLKPEFID 242 Query: 774 IINAK 788 IINA+ Sbjct: 243 IINAR 247
>sp|P00939|TPIS_RABIT Triosephosphate isomerase (TIM) (Triose-phosphate isomerase) Length = 248 Score = 358 bits (919), Expect = 1e-98 Identities = 173/244 (70%), Positives = 199/244 (81%) Frame = +3 Query: 57 RKFFVGGNWKMNGDKDELTKICNSLMNAKLHETTEVVVAPPSIYLDFVRQKLPAKIGVSA 236 RKFFVGGNWKMNG K L ++ +L AK+ TEVV APP+ Y+DF RQKL KI V+A Sbjct: 4 RKFFVGGNWKMNGRKKNLGELITTLNAAKVPADTEVVCAPPTAYIDFARQKLDPKIAVAA 63 Query: 237 QNCYKIPKGAFTGEISPAMIKDNMCDWVILGHSERRHVFGESDHLIGEKVKHALESGLSI 416 QNCYK+ GAFTGEISP MIKD WV+LGHSERRHVFGESD LIG+KV HAL GL + Sbjct: 64 QNCYKVTNGAFTGEISPGMIKDCGATWVVLGHSERRHVFGESDELIGQKVAHALSEGLGV 123 Query: 417 IPCIGEKLDERESNKTNEVCFRQLKEIAANIKDWTRVVIAYEPVWAIGTGKTASPAQAQE 596 I CIGEKLDERE+ T +V F Q K IA N+KDW++VV+AYEPVWAIGTGKTA+P QAQE Sbjct: 124 IACIGEKLDEREAGITEKVVFEQTKVIADNVKDWSKVVLAYEPVWAIGTGKTATPQQAQE 183 Query: 597 VHKCVRDWLKTNVSEEVAKTTRILYGGSVTSGNCKELASQPDVDGFLVGGASLKPEFIDI 776 VH+ +R WLK+NVS+ VA++TRI+YGGSVT CKELASQPDVDGFLVGGASLKPEF+DI Sbjct: 184 VHEKLRGWLKSNVSDAVAQSTRIIYGGSVTGATCKELASQPDVDGFLVGGASLKPEFVDI 243 Query: 777 INAK 788 INAK Sbjct: 244 INAK 247
>sp|P48500|TPIS_RAT Triosephosphate isomerase (TIM) (Triose-phosphate isomerase) Length = 249 Score = 357 bits (916), Expect = 3e-98 Identities = 175/248 (70%), Positives = 198/248 (79%) Frame = +3 Query: 45 MSNQRKFFVGGNWKMNGDKDELTKICNSLMNAKLHETTEVVVAPPSIYLDFVRQKLPAKI 224 M+ RKFFVGGNWKMNG K L ++ +L AKL TEVV APP+ Y+DF RQKL KI Sbjct: 1 MAPSRKFFVGGNWKMNGRKKCLGELICTLNAAKLPADTEVVCAPPTAYIDFARQKLDPKI 60 Query: 225 GVSAQNCYKIPKGAFTGEISPAMIKDNMCDWVILGHSERRHVFGESDHLIGEKVKHALES 404 V+AQNCYK+ GAFTGEISP MIKD WV+LGHSERRH+FGESD LIG+KV HAL Sbjct: 61 AVAAQNCYKVTNGAFTGEISPGMIKDLGATWVVLGHSERRHIFGESDELIGQKVNHALSE 120 Query: 405 GLSIIPCIGEKLDERESNKTNEVCFRQLKEIAANIKDWTRVVIAYEPVWAIGTGKTASPA 584 GL +I CIGEKLDERE+ T +V F Q K IA N+KDW +VV+AYEPVWAIGTGKTA+P Sbjct: 121 GLEVIACIGEKLDEREAGITEKVVFEQTKAIADNVKDWCKVVLAYEPVWAIGTGKTATPQ 180 Query: 585 QAQEVHKCVRDWLKTNVSEEVAKTTRILYGGSVTSGNCKELASQPDVDGFLVGGASLKPE 764 QAQEVH+ +R WLK NVSE VA+ TRI+YGGSVT CKELASQPDVDGFLVGGASLKPE Sbjct: 181 QAQEVHEKLRGWLKCNVSEGVAQCTRIIYGGSVTGATCKELASQPDVDGFLVGGASLKPE 240 Query: 765 FIDIINAK 788 F+DIINAK Sbjct: 241 FVDIINAK 248
>sp|P17751|TPIS_MOUSE Triosephosphate isomerase (TIM) (Triose-phosphate isomerase) Length = 249 Score = 353 bits (905), Expect = 5e-97 Identities = 172/248 (69%), Positives = 200/248 (80%) Frame = +3 Query: 45 MSNQRKFFVGGNWKMNGDKDELTKICNSLMNAKLHETTEVVVAPPSIYLDFVRQKLPAKI 224 M+ RKFFVGGNWKMNG K L ++ +L A + TEVV APP+ Y+DF RQKL KI Sbjct: 1 MAPTRKFFVGGNWKMNGRKKCLGELICTLNAANVPAGTEVVCAPPTAYIDFARQKLDPKI 60 Query: 225 GVSAQNCYKIPKGAFTGEISPAMIKDNMCDWVILGHSERRHVFGESDHLIGEKVKHALES 404 V+AQNCYK+ GAFTGEISP MIKD WV+LGHSERRHVFGESD LIG+KV HAL Sbjct: 61 AVAAQNCYKVTNGAFTGEISPGMIKDLGATWVVLGHSERRHVFGESDELIGQKVSHALAE 120 Query: 405 GLSIIPCIGEKLDERESNKTNEVCFRQLKEIAANIKDWTRVVIAYEPVWAIGTGKTASPA 584 GL +I CIGEKLDERE+ T +V F Q K IA N+KDW++VV+AYEPVWAIGTGKTA+P Sbjct: 121 GLGVIACIGEKLDEREAGITEKVVFEQTKVIADNVKDWSKVVLAYEPVWAIGTGKTATPQ 180 Query: 585 QAQEVHKCVRDWLKTNVSEEVAKTTRILYGGSVTSGNCKELASQPDVDGFLVGGASLKPE 764 QAQEVH+ +R WLK+NV++ VA++TRI+YGGSVT CKELASQPDVDGFLVGGASLKPE Sbjct: 181 QAQEVHEKLRGWLKSNVNDGVAQSTRIIYGGSVTGATCKELASQPDVDGFLVGGASLKPE 240 Query: 765 FIDIINAK 788 F+DIINAK Sbjct: 241 FVDIINAK 248
>sp|P48501|TPIS_SCHMA Triosephosphate isomerase (TIM) (Triose-phosphate isomerase) Length = 253 Score = 339 bits (870), Expect = 6e-93 Identities = 166/251 (66%), Positives = 193/251 (76%), Gaps = 3/251 (1%) Frame = +3 Query: 45 MSNQRKFFVGGNWKMNGDKDELTKICNSLMNAKLHETTEVVVAPPSIYLDFVRQKLPAKI 224 MS RKFFVGGNWKMNG +D+ K+ L A + TEV++APPS++L +R+ L +I Sbjct: 1 MSGSRKFFVGGNWKMNGSRDDNDKLLKLLSEAHFDDNTEVLIAPPSVFLHEIRKSLKKEI 60 Query: 225 GVSAQNCYKIPKGAFTGEISPAMIKDNMCDWVILGHSERRHVFGESDHLIGEKVKHALES 404 V+AQNCYK+ KGAFTGEISPAMI+D CDWVILGHSERR++FGESD LI EKV+HAL Sbjct: 61 HVAAQNCYKVSKGAFTGEISPAMIRDIGCDWVILGHSERRNIFGESDELIAEKVQHALAE 120 Query: 405 GLSIIPCIGEKLDERESNKTNEVCFRQLKEIAANIK---DWTRVVIAYEPVWAIGTGKTA 575 GLS+I CIGE L ERESNKT EVC RQLK IA IK +W RVV+AYEPVWAIGTGK A Sbjct: 121 GLSVIACIGETLSERESNKTEEVCVRQLKAIANKIKSADEWKRVVVAYEPVWAIGTGKVA 180 Query: 576 SPAQAQEVHKCVRDWLKTNVSEEVAKTTRILYGGSVTSGNCKELASQPDVDGFLVGGASL 755 +P QAQEVH +R W KTN V + RI+YGGSVT+ NCKELA Q DVDGFLVGGASL Sbjct: 181 TPQQAQEVHNFLRKWFKTNAPNGVDEKIRIIYGGSVTAANCKELAQQHDVDGFLVGGASL 240 Query: 756 KPEFIDIINAK 788 KPEF +I A+ Sbjct: 241 KPEFTEICKAR 251
>sp|P00941|TPIS_LATCH Triosephosphate isomerase (TIM) (Triose-phosphate isomerase) Length = 247 Score = 337 bits (864), Expect = 3e-92 Identities = 163/239 (68%), Positives = 189/239 (79%) Frame = +3 Query: 57 RKFFVGGNWKMNGDKDELTKICNSLMNAKLHETTEVVVAPPSIYLDFVRQKLPAKIGVSA 236 RKFFVGGNWKMNGDK L ++ +L AK+ T E+V APP YLDF R K+ K GV+A Sbjct: 3 RKFFVGGNWKMNGDKKSLGELIQTLNAAKVPFTGEIVCAPPEAYLDFARLKVDPKFGVAA 62 Query: 237 QNCYKIPKGAFTGEISPAMIKDNMCDWVILGHSERRHVFGESDHLIGEKVKHALESGLSI 416 QNCYK+ KGAFTGEISPAMIKD WVILGHSERRHVFGESD LIG+KV HAL GL + Sbjct: 63 QNCYKVSKGAFTGEISPAMIKDCGVTWVILGHSERRHVFGESDELIGQKVSHALSEGLGV 122 Query: 417 IPCIGEKLDERESNKTNEVCFRQLKEIAANIKDWTRVVIAYEPVWAIGTGKTASPAQAQE 596 + CIGEKLDERE+ T V F + IA ++KDW++VV+AYEPVWAIGTGKTASP Q+QE Sbjct: 123 VACIGEKLDEREAGITEGVVFEVTEVIADDVKDWSKVVLAYEPVWAIGTGKTASPQQSQE 182 Query: 597 VHKCVRDWLKTNVSEEVAKTTRILYGGSVTSGNCKELASQPDVDGFLVGGASLKPEFID 773 +H +R WLK NVSE VA + RI+YGGSVT CKELAS+PDVDGFLVGGASLKPEF++ Sbjct: 183 LHGKLRKWLKENVSETVADSVRIIYGGSVTGATCKELASEPDVDGFLVGGASLKPEFVE 241
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 94,739,702
Number of Sequences: 369166
Number of extensions: 1797874
Number of successful extensions: 5182
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 4672
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 4814
length of database: 68,354,980
effective HSP length: 111
effective length of database: 47,849,395
effective search space used: 10048372950
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail