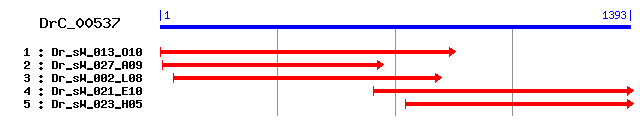
DrC_00537
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00537 (1391 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q38924|PPAF_ARATH Iron(III)-zinc(II) purple acid phospha... 121 6e-27 sp||Q687E1_2 [Segment 2 of 2] Nucleotide pyrophosphatase/ph... 111 6e-24 sp|P80366|PPAF_PHAVU Iron(III)-zinc(II) purple acid phospha... 110 1e-23 sp|Q12546|PPA_ASPFI Acid phosphatase precursor (pH 6-optimu... 78 7e-14 sp|Q50644|YP77_MYCTU Hypothetical protein Rv2577/MT2654 54 1e-06 sp|Q05205|PPB_LYSEN Alkaline phosphatase precursor (APASE) 41 0.007 sp|P41586|PACR_HUMAN Pituitary adenylate cyclase-activating... 34 1.2 sp|P0AC44|DHSD_ECOLI Succinate dehydrogenase hydrophobic me... 33 2.0 sp|Q49161|ACDA1_METMA Acetyl-CoA decarbonylase/synthase com... 33 2.0 sp|P19274|VTPX_TTV1 Viral protein TPX 32 3.4
>sp|Q38924|PPAF_ARATH Iron(III)-zinc(II) purple acid phosphatase precursor (PAP) Length = 469 Score = 121 bits (303), Expect = 6e-27 Identities = 115/437 (26%), Positives = 183/437 (41%), Gaps = 24/437 (5%) Frame = +3 Query: 57 YNQPEQIHLSIGSNI-NXXXXXXXXXXXXNESIVIYGPDKSMNLKQ-NGTVD--KFVDGG 224 +N P+Q+H++ G++ N V Y + + KQ TV+ +F + Sbjct: 57 HNSPQQVHVTQGNHEGNGVIISWVTPVKPGSKTVQYWCENEKSRKQAEATVNTYRFFNYT 116 Query: 225 SEKRQIFMHRVILPNLLPFTQYSYTVGSRFGWSEIHTFKTFPP--GSNWPTRYVVVGDMG 398 S ++H ++ +L T+Y Y +GS WS F PP G + P + ++GD+G Sbjct: 117 SG----YIHHCLIDDLEFDTKYYYEIGSG-KWSRRFWF-FIPPKSGPDVPYTFGLIGDLG 170 Query: 399 NLNARSLAAIQRETEKGFYDMVLHVGDFAY--NMDSYNAQTGDQFMRQIQPVAAQLPYMT 572 + E G VL VGD +Y +++ D + R ++ A P++ Sbjct: 171 QTYDSNSTLSHYEMNPGKGQAVLFVGDLSYADRYPNHDNNRWDTWGRFVERSVAYQPWIW 230 Query: 573 VPGNHE--------EAYNFSNYRRRFTMP---GGDGEGQYFSFNVGYAHIIGFSSEFYYF 719 GNHE E F + R+ P G ++S A+II S + Sbjct: 231 TAGNHEIDFVPDIGEIEPFKPFMNRYHTPHKASGSISPLWYSIKRASAYIIVMSC----Y 286 Query: 720 TNYGFKQIENQYNWLLNDLKEANKPENRAVRPWIIAMGHRPMYCSNSDDGQHCENLINSI 899 ++YG QY WL +L+ N+ E PW+I + H P Y S Sbjct: 287 SSYGI--YTPQYKWLEKELQGVNRTET----PWLIVLVHSPFYSSY-------------- 326 Query: 900 RVGMPISVHRY--GSMFVFGLEKLFYDYGVDLIFGAHEHSYERMWPVYNL--KVCNGTEA 1067 VH Y G E+ F Y VD++F H H+YER V N+ + NG Sbjct: 327 -------VHHYMEGETLRVMYEQWFVKYKVDVVFAGHVHAYERSERVSNIAYNIVNGLCE 379 Query: 1068 PYTDPPAPVHIVTGSAGNQEG-EDPFYANKPDWSAFHSSDYGYTRMNIMNSSHMYLEQVS 1244 P +D AP++I G GN EG +P +SAF + +G+ + I N +H Y Sbjct: 380 PISDESAPIYITIGDGGNSEGLLTDMMQPQPKYSAFREASFGHGLLEIKNRTHAYFSWNR 439 Query: 1245 DDLGGAVIDKILYIRNK 1295 + G AV +++ N+ Sbjct: 440 NQDGNAVAADSVWLLNR 456
>sp||Q687E1_2 [Segment 2 of 2] Nucleotide pyrophosphatase/phosphodiesterase Length = 350 Score = 111 bits (277), Expect = 6e-24 Identities = 99/360 (27%), Positives = 154/360 (42%), Gaps = 26/360 (7%) Frame = +3 Query: 318 WSEIHTFKTFP-PGSNWPTRYVVVGDMG-----------NLNARSLAAIQRETEK-GFYD 458 W++ +TF+ P PG N R +V GDMG N SL R E YD Sbjct: 7 WAKPYTFRAPPTPGQNSLQRIIVFGDMGKAERDGSNEFANYQPGSLNTTDRLIEDLDNYD 66 Query: 459 MVLHVGDFAYNMDSYNAQTGDQFMRQIQPVAAQLPYMTVPGNHEEAYNFSNYRRRFTMPG 638 +V H+GD Y + Y +Q DQF Q+ P++A+ PYM GNHE ++ N F + Sbjct: 67 IVFHIGDMPY-ANGYLSQW-DQFTAQVAPISAKKPYMVASGNHER--DWPNTGGFFDVKD 122 Query: 639 GDGEGQYFSFNVGYAHIIGFSSEFYYFTNYGFKQI---ENQYNW-----LLNDLKEANKP 794 GE + + Y + + F+Y +YG + +++++W ++E Sbjct: 123 SGGECGVPAETM-YYYPAENRANFWYKVDYGMFRFCVGDSEHDWREGTPQYKFIEECLST 181 Query: 795 ENRAVRPWIIAMGHRPM-YCSNS---DDGQHCENLINSIRVGMPISVHRYGSMFVFGLEK 962 +R +PW+I HR + Y SNS D G E L+K Sbjct: 182 VDRKHQPWLIFTAHRVLGYSSNSWYADQGSFEEPEGRE------------------SLQK 223 Query: 963 LFYDYGVDLIFGAHEHSYERMWPVYNLKVCNGTEAPYTDP-PAPVHIVTGSAGNQEGEDP 1139 L+ Y VD+ + H H+YER P+Y + N + Y+ + +V G G+ Sbjct: 224 LWQRYRVDIAYFGHVHNYERTCPLYQSQCVNADKTHYSGTMNGTIFVVAGGGGSHLSS-- 281 Query: 1140 FYANKPDWSAFHSSDYGYTRMNIMNSSHMYLEQVSDDLGGAVIDKILYIRNKHGQIFNCV 1319 + P WS F DYG+T++ N S + E + G V D I + + +CV Sbjct: 282 YTTAIPKWSIFRDHDYGFTKLTAFNHSSLLFEYMKSS-DGKVYDSFT-IHRDYRDVLSCV 339
>sp|P80366|PPAF_PHAVU Iron(III)-zinc(II) purple acid phosphatase (PAP) Length = 432 Score = 110 bits (275), Expect = 1e-23 Identities = 116/440 (26%), Positives = 181/440 (41%), Gaps = 26/440 (5%) Frame = +3 Query: 57 YNQPEQIHLSIGSNINXXXXXXXXXXXX-NESIVIYGPDKSMN---LKQNGTVDKFVDGG 224 YN P+Q+H++ G + S V Y +K+ K + +F + Sbjct: 24 YNAPQQVHITQGDLVGRAMIISWVTMDEPGSSAVRYWSEKNGRKRIAKGKMSTYRFFNYS 83 Query: 225 SEKRQIFMHRVILPNLLPFTQYSYTVGSRFGWSEIHTFKTFPP-GSNWPTRYVVVGDMGN 401 S F+H + L T+Y Y VG R + +F T P G + P + ++GD+G Sbjct: 84 SG----FIHHTTIRKLKYNTKYYYEVGLR-NTTRRFSFITPPQTGLDVPYTFGLIGDLGQ 138 Query: 402 L--NARSLAAIQRETEKGFYDMVLHVGDFAY-----NMDSYNAQTGDQFMRQIQPVAAQL 560 + +L+ + +KG VL VGD +Y N D+ T +F + A Sbjct: 139 SFDSNTTLSHYELSPKKG--QTVLFVGDLSYADRYPNHDNVRWDTWGRFTERS---VAYQ 193 Query: 561 PYMTVPGNHE--------EAYNFSNYRRRFTMPGGDGEGQ---YFSFNVGYAHIIGFSSE 707 P++ GNHE E F + R+ +P + ++S AHII SS Sbjct: 194 PWIWTAGNHEIEFAPEINETEPFKPFSYRYHVPYEASQSTSPFWYSIKRASAHIIVLSSH 253 Query: 708 FYYFTNYGFKQIENQYNWLLNDLKEANKPENRAVRPWIIAMGHRPMYCSNSDDGQHCENL 887 Y QY WL +L++ + E PW+I + H P+Y NS + E Sbjct: 254 IAYGRG------TPQYTWLKKELRKVKRSET----PWLIVLMHSPLY--NSYNHHFME-- 299 Query: 888 INSIRVGMPISVHRYGSMFVFGLEKLFYDYGVDLIFGAHEHSYERMWPVYNL--KVCNGT 1061 G E F Y VD++F H H+YER V N+ K+ +G Sbjct: 300 ---------------GEAMRTKFEAWFVKYKVDVVFAGHVHAYERSERVSNIAYKITDGL 344 Query: 1062 EAPYTDPPAPVHIVTGSAGNQEGEDP-FYANKPDWSAFHSSDYGYTRMNIMNSSHMYLEQ 1238 P D APV+I G AGN D +P++SAF + +G+ +I N +H + Sbjct: 345 CTPVKDQSAPVYITIGDAGNYGVIDSNMIQPQPEYSAFREASFGHGMFDIKNRTHAHFSW 404 Query: 1239 VSDDLGGAVIDKILYIRNKH 1298 + G AV ++ N+H Sbjct: 405 NRNQDGVAVEADSVWFFNRH 424
>sp|Q12546|PPA_ASPFI Acid phosphatase precursor (pH 6-optimum acid phosphatase) (APase6) Length = 614 Score = 77.8 bits (190), Expect = 7e-14 Identities = 108/496 (21%), Positives = 162/496 (32%), Gaps = 144/496 (29%) Frame = +3 Query: 243 FMHRVILPNLLPFTQYSYTVGSRFGW--SEIHTFKTF-PPGSNWPTRYVVVGDMGNLNAR 413 F H V + L P T Y Y + + G SE+ +FKT P G V+ DMG NA Sbjct: 138 FFHEVSIDGLEPDTTYYYQIPAANGTTQSEVLSFKTSRPAGHPGSFSVAVLNDMGYTNAH 197 Query: 414 SL--AAIQRETEKGFYDMVLHVGDFAYNMD--------------SYNAQTG--------- 518 ++ TE + H GD +Y D YN + Sbjct: 198 GTHKQLVKAATEGTAF--AWHGGDLSYADDWYSGILACADDWPVCYNGTSSTLPGGGPLP 255 Query: 519 -------------------------------DQFMRQIQPVAAQLPYMTVPGNHEE---- 593 D + + + V ++PYM +PGNHE Sbjct: 256 EEYKKPLPAGEIPDQGGPQGGDMSVLYESNWDLWQQWLNNVTLKIPYMVLPGNHEASCAE 315 Query: 594 ---------AY-------------------------NFSNYRRRFTMPGGDGEGQ---YF 662 AY NF+ Y+ RF MPG + G ++ Sbjct: 316 FDGPHNILTAYLNDDIANGTAPTDNLTYYSCPPSQRNFTAYQHRFRMPGPETGGVGNFWY 375 Query: 663 SFNVGYAHIIGFSSEFYYFTNYGFKQIEN------------------------------- 749 SF+ G AH + E + + + E+ Sbjct: 376 SFDYGLAHFVSIDGETDFANSPEWNFAEDVTGNETLPSESETFITDSGPFGNVNGSVHET 435 Query: 750 ----QYNWLLNDLKEANKPENRAVRPWIIAMGHRPMYCSNSDDGQHCENLINSIRVGMPI 917 Q++WL DL + + R+ PW+I M HRPMY S Sbjct: 436 KSYEQWHWLQQDLAKVD----RSKTPWVIVMSHRPMYSS--------------------- 470 Query: 918 SVHRYGSMFVFGLEKLFYDYGVDLIFGAHEHSYERMWP------VYNLKVCNGTEAPYTD 1079 + Y E L YGVD H H YER++P + + N + Sbjct: 471 AYSSYQLHVREAFEGLLLKYGVDAYLSGHIHWYERLYPLGANGTIDTAAIVNNNTYYAHN 530 Query: 1080 PPAPVHIVTGSAGNQEGEDPFYANKPDWSAFHSSD---YGYTRMNIMNSSHMYLEQVSDD 1250 + HI+ G AGN E F + + D YG++++ I N + + E + D Sbjct: 531 GKSITHIINGMAGNIESHSEFSDGEGLTNITALLDKVHYGFSKLTIFNETALKWELIRGD 590 Query: 1251 LGGAVIDKILYIRNKH 1298 G V D + ++ H Sbjct: 591 -DGTVGDSLTLLKPSH 605
>sp|Q50644|YP77_MYCTU Hypothetical protein Rv2577/MT2654 Length = 529 Score = 53.9 bits (128), Expect = 1e-06 Identities = 88/397 (22%), Positives = 130/397 (32%), Gaps = 73/397 (18%) Frame = +3 Query: 237 QIFMHRVILPNLLPFTQYSYTVGSRFGWSEIHTFKTFPPGSNWPTRYVVVGDMG------ 398 ++ ++ L NL P T Y Y E+ T +T P G P R+ GD Sbjct: 123 EVRVNHAHLTNLTPDTDYVYAAVHDGTTPELGTARTAPSGRK-PLRFTSFGDQSTPALGR 181 Query: 399 ---------NLNARSLAAIQRETEKGFYDMVLHVGDFAY-NMDSYNAQTGDQFMRQIQPV 548 N+ + I E+ L GD Y N+ +T + Sbjct: 182 LADGRYVSDNIGSPFAGDITIAIERIAPLFNLINGDLCYANLAQDRIRTWSDWFDNNTRS 241 Query: 549 AAQLPYMTVPGNHEEAYN-----FSNYRRRFTMPGGDGE----GQYFSFNVGYAHIIGFS 701 A P+M GNHE + Y+ F +P G ++SF G +I Sbjct: 242 ARYRPWMPAAGNHENEVGNGPIGYDAYQTYFAVPDSGSSPQLRGLWYSFTAGSVRVISLH 301 Query: 702 SEFYYFTNYGFKQIEN-----QYNWLLNDLKEANKPENRAVRPWIIAMGHRPMYCSNSDD 866 ++ + + G + Q WL +L A + W++ H+ S +DD Sbjct: 302 NDDVCYQDGGNSYVRGYSGGEQRRWLQAELANARRDSEI---DWVVVCMHQTAI-STADD 357 Query: 867 GQHCENLINSIRVGMPISVHRYGSMFVFGLEKLFYDYGVDLIFGAHEHSYERMWPVYNLK 1046 + I + +P LF Y VDL+ HEH YER P L+ Sbjct: 358 NNGADLGIR--QEWLP----------------LFDQYQVDLVVCGHEHHYERSHP---LR 396 Query: 1047 VCNGTEAPYTDPPAP--------------VHIVTGSAGNQEGE----------------- 1133 GT+ T P P VH+V G G + Sbjct: 397 GALGTD---TRTPIPVDTRSDLIDSTRGTVHLVIGGGGTSKPTNALLFPQPRCQVITGVG 453 Query: 1134 --DPFYANKPD--------WSAFHSSD--YGYTRMNI 1208 DP KP WSAF D YG+ ++ Sbjct: 454 DFDPAIRRKPSIFVLEDAPWSAFRDRDNPYGFVAFDV 490
>sp|Q05205|PPB_LYSEN Alkaline phosphatase precursor (APASE) Length = 539 Score = 41.2 bits (95), Expect = 0.007 Identities = 52/227 (22%), Positives = 84/227 (37%), Gaps = 12/227 (5%) Frame = +3 Query: 378 VVVGDMGNLNARSLAAIQRETEKGFYDMVLHVGD---FAYNMDSYNAQTGDQFMRQIQPV 548 VVV G++ S A Q G D+++ + F ++YN+ T ++ + P Sbjct: 151 VVVAGAGDICDTSGNACQ-----GTSDLIVSINPTAVFTAGDNAYNSGTLSEYNSRYAPT 205 Query: 549 AAQLPYMTVP--GNHEEA-------YNFSNYRRRFTMPGGDGEGQYFSFNVGYAHIIGFS 701 + +T P GNH+ + +++ N T P GD Y+S++VG H + + Sbjct: 206 WGRFKALTSPSPGNHDYSTTGAKGYFDYFNGSGNQTGPAGDRSKGYYSWDVGDWHFVSLN 265 Query: 702 SEFYYFTNYGFKQIENQYNWLLNDLKEANKPENRAVRPWIIAMGHRPMYCSNSDDGQHCE 881 T G + Q +WL DL K P A H P+ S G Sbjct: 266 ------TMSGGTVAQAQIDWLKADLAANTK-------PCTAAYFHHPLLSRGSYSG---- 308 Query: 882 NLINSIRVGMPISVHRYGSMFVFGLEKLFYDYGVDLIFGAHEHSYER 1022 Y + F Y DL+ H+H+Y+R Sbjct: 309 ----------------YSQVKPFW--DALYAAKADLVLVGHDHNYQR 337
>sp|P41586|PACR_HUMAN Pituitary adenylate cyclase-activating polypeptide type I receptor precursor (PACAP type I receptor) (PACAP-R-1) Length = 468 Score = 33.9 bits (76), Expect = 1.2 Identities = 22/67 (32%), Positives = 32/67 (47%), Gaps = 4/67 (5%) Frame = +3 Query: 336 FKTFPPGSNWPTRYVVVGDMGNLNARSLA---AIQRE-TEKGFYDMVLHVGDFAYNMDSY 503 F+ F P W T + D G+ N+ L+ + R TE G+ + H D A D Y Sbjct: 81 FRIFNPDQVWETETIGESDFGDSNSLDLSDMGVVSRNCTEDGWSEPFPHYFD-ACGFDEY 139 Query: 504 NAQTGDQ 524 ++TGDQ Sbjct: 140 ESETGDQ 146
>sp|P0AC44|DHSD_ECOLI Succinate dehydrogenase hydrophobic membrane anchor protein sp|P0AC46|DHSD_SHIFL Succinate dehydrogenase hydrophobic membrane anchor protein sp|P0AC45|DHSD_ECOL6 Succinate dehydrogenase hydrophobic membrane anchor protein Length = 115 Score = 33.1 bits (74), Expect = 2.0 Identities = 13/34 (38%), Positives = 23/34 (67%), Gaps = 2/34 (5%) Frame = +2 Query: 242 FYASSYTTEF--VAIYTIQLHCWIKIWLVRNSYI 337 F+AS++T F +A+++I +H WI +W V Y+ Sbjct: 51 FFASAFTKVFTLLALFSILIHAWIGMWQVLTDYV 84
>sp|Q49161|ACDA1_METMA Acetyl-CoA decarbonylase/synthase complex alpha subunit 1 (ACDS complex alpha subunit 1) (ACDS complex carbon monoxide dehydrogenase 1) (ACDS CODH 1) Length = 806 Score = 33.1 bits (74), Expect = 2.0 Identities = 21/61 (34%), Positives = 28/61 (45%), Gaps = 10/61 (16%) Frame = +3 Query: 975 YGVDLIFGAHEHSYER----------MWPVYNLKVCNGTEAPYTDPPAPVHIVTGSAGNQ 1124 YG+ + GAH Y R W VY+ + NG E P PPAP ++T + Q Sbjct: 648 YGIPAVLGAHSSKYRRALIAKNYDESKWKVYDAR--NGEEMPI--PPAPEFLLTTAETWQ 703 Query: 1125 E 1127 E Sbjct: 704 E 704
>sp|P19274|VTPX_TTV1 Viral protein TPX Length = 360 Score = 32.3 bits (72), Expect = 3.4 Identities = 14/36 (38%), Positives = 21/36 (58%) Frame = -2 Query: 361 LEPGGKVLNV*ISDQPNLDPTV*LYCVNGNKFGSIT 254 L PG + S PN+DPT+ LY NG+ + ++T Sbjct: 59 LSPGQSITITASSGTPNIDPTIALYYNNGSSYSNLT 94
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 167,326,526
Number of Sequences: 369166
Number of extensions: 3698885
Number of successful extensions: 8549
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 8125
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 8533
length of database: 68,354,980
effective HSP length: 114
effective length of database: 47,295,190
effective search space used: 16506021310
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail