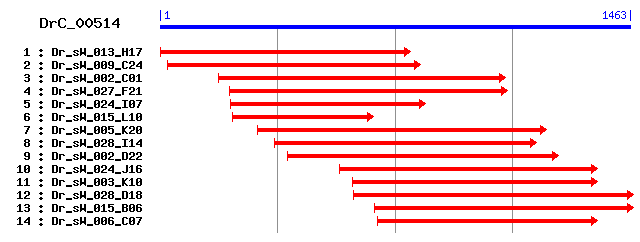
DrC_00514
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00514 (1462 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P11833|TBB_PARLI Tubulin beta chain (Beta tubulin) 806 0.0 sp|P68372|TBBX_MOUSE Tubulin beta-? chain >gi|55977480|sp|P... 806 0.0 sp|P30883|TBB4_XENLA Tubulin beta-4 chain 804 0.0 sp|P02554|TBB_PIG Tubulin beta chain 798 0.0 sp|P32882|TBB2_CHICK Tubulin beta-2 chain (Beta-tubulin cla... 798 0.0 sp|Q7JJU6|TBB2_PANTR Tubulin beta-2 chain >gi|56754798|sp|P... 798 0.0 sp|P09206|TBB3_CHICK Tubulin beta-3 chain (Beta-tubulin cla... 797 0.0 sp|Q91240|TBB_PSEAM Tubulin beta chain (Beta tubulin) 797 0.0 sp|O17449|TBB1_MANSE Tubulin beta-1 chain (Beta-1 tubulin) 795 0.0 sp|P09203|TBB1_CHICK Tubulin beta-1 chain (Beta-tubulin cla... 795 0.0
>sp|P11833|TBB_PARLI Tubulin beta chain (Beta tubulin) Length = 447 Score = 806 bits (2082), Expect = 0.0 Identities = 392/418 (93%), Positives = 399/418 (95%) Frame = +2 Query: 2 GNQIGAKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEATGGKYVPRAVLIDLEPGT 181 GNQIGAKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEATGGKYVPRAVL+DLEPGT Sbjct: 13 GNQIGAKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEATGGKYVPRAVLVDLEPGT 72 Query: 182 MDSVRAGPFGQLFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDIVRKEAESCDCLQG 361 MDSVR+GPFGQ+FRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLD+VRKEAESCDCLQG Sbjct: 73 MDSVRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDVVRKEAESCDCLQG 132 Query: 362 FQXXXXXXXXXXXXXXXXXXXKIREEYPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQL 541 FQ KIREEYPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQL Sbjct: 133 FQLTHSLGGGTGSGMGTLLISKIREEYPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQL 192 Query: 542 VENTDETFCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRK 721 VENTDET+CIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRK Sbjct: 193 VENTDETYCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRK 252 Query: 722 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMMAACDPRHGRYLT 901 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMMAACDPRHGRYLT Sbjct: 253 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMMAACDPRHGRYLT 312 Query: 902 VAALFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMSATFIGNST 1081 VAA+FRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMSATFIGNST Sbjct: 313 VAAIFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMSATFIGNST 372 Query: 1082 AIQELFKRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA 1255 AIQELFKR+SEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA Sbjct: 373 AIQELFKRISEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA 430
>sp|P68372|TBBX_MOUSE Tubulin beta-? chain sp|P68371|TBBX_HUMAN Tubulin beta-? chain (Tubulin beta-2 chain) Length = 445 Score = 806 bits (2081), Expect = 0.0 Identities = 392/418 (93%), Positives = 399/418 (95%) Frame = +2 Query: 2 GNQIGAKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEATGGKYVPRAVLIDLEPGT 181 GNQIGAKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEATGGKYVPRAVL+DLEPGT Sbjct: 13 GNQIGAKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEATGGKYVPRAVLVDLEPGT 72 Query: 182 MDSVRAGPFGQLFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDIVRKEAESCDCLQG 361 MDSVR+GPFGQ+FRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLD+VRKEAESCDCLQG Sbjct: 73 MDSVRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDVVRKEAESCDCLQG 132 Query: 362 FQXXXXXXXXXXXXXXXXXXXKIREEYPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQL 541 FQ KIREEYPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQL Sbjct: 133 FQLTHSLGGGTGSGMGTLLISKIREEYPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQL 192 Query: 542 VENTDETFCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRK 721 VENTDET+CIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRK Sbjct: 193 VENTDETYCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRK 252 Query: 722 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMMAACDPRHGRYLT 901 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMMAACDPRHGRYLT Sbjct: 253 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMMAACDPRHGRYLT 312 Query: 902 VAALFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMSATFIGNST 1081 VAA+FRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMSATFIGNST Sbjct: 313 VAAVFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMSATFIGNST 372 Query: 1082 AIQELFKRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA 1255 AIQELFKR+SEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA Sbjct: 373 AIQELFKRISEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA 430
>sp|P30883|TBB4_XENLA Tubulin beta-4 chain Length = 445 Score = 804 bits (2077), Expect = 0.0 Identities = 391/418 (93%), Positives = 398/418 (95%) Frame = +2 Query: 2 GNQIGAKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEATGGKYVPRAVLIDLEPGT 181 GNQIGAKFWEVISDEHGIDPTG YHGDSDLQLERINVYYNEATGGKYVPRAVL+DLEPGT Sbjct: 13 GNQIGAKFWEVISDEHGIDPTGAYHGDSDLQLERINVYYNEATGGKYVPRAVLVDLEPGT 72 Query: 182 MDSVRAGPFGQLFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDIVRKEAESCDCLQG 361 MDSVR+GPFGQ+FRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLD+VRKEAESCDCLQG Sbjct: 73 MDSVRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDVVRKEAESCDCLQG 132 Query: 362 FQXXXXXXXXXXXXXXXXXXXKIREEYPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQL 541 FQ KIREEYPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQL Sbjct: 133 FQLTHSLGGGTGSGMGTLLISKIREEYPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQL 192 Query: 542 VENTDETFCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRK 721 VENTDET+CIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRK Sbjct: 193 VENTDETYCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRK 252 Query: 722 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMMAACDPRHGRYLT 901 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMMAACDPRHGRYLT Sbjct: 253 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMMAACDPRHGRYLT 312 Query: 902 VAALFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMSATFIGNST 1081 VAA+FRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMSATFIGNST Sbjct: 313 VAAIFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMSATFIGNST 372 Query: 1082 AIQELFKRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA 1255 AIQELFKR+SEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA Sbjct: 373 AIQELFKRISEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA 430
>sp|P02554|TBB_PIG Tubulin beta chain Length = 445 Score = 798 bits (2062), Expect = 0.0 Identities = 387/418 (92%), Positives = 397/418 (94%) Frame = +2 Query: 2 GNQIGAKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEATGGKYVPRAVLIDLEPGT 181 GNQIGAKFWEVISDEHGIDPTG+YHGDSDLQLERINVYYNEA G KYVPRA+L+DLEPGT Sbjct: 13 GNQIGAKFWEVISDEHGIDPTGSYHGDSDLQLERINVYYNEAAGNKYVPRAILVDLEPGT 72 Query: 182 MDSVRAGPFGQLFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDIVRKEAESCDCLQG 361 MDSVR+GPFGQ+FRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLD+VRKE+ESCDCLQG Sbjct: 73 MDSVRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDVVRKESESCDCLQG 132 Query: 362 FQXXXXXXXXXXXXXXXXXXXKIREEYPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQL 541 FQ KIREEYPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQL Sbjct: 133 FQLTHSLGGGTGSGMGTLLISKIREEYPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQL 192 Query: 542 VENTDETFCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRK 721 VENTDET+CIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRK Sbjct: 193 VENTDETYCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRK 252 Query: 722 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMMAACDPRHGRYLT 901 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMMAACDPRHGRYLT Sbjct: 253 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMMAACDPRHGRYLT 312 Query: 902 VAALFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMSATFIGNST 1081 VAA+FRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMSATFIGNST Sbjct: 313 VAAVFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMSATFIGNST 372 Query: 1082 AIQELFKRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA 1255 AIQELFKR+SEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA Sbjct: 373 AIQELFKRISEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA 430
>sp|P32882|TBB2_CHICK Tubulin beta-2 chain (Beta-tubulin class-II) Length = 445 Score = 798 bits (2062), Expect = 0.0 Identities = 386/418 (92%), Positives = 398/418 (95%) Frame = +2 Query: 2 GNQIGAKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEATGGKYVPRAVLIDLEPGT 181 GNQIGAKFWEVISDEHGIDPTG+YHGDSDLQLERINVYYNEATG KYVPRA+L+DLEPGT Sbjct: 13 GNQIGAKFWEVISDEHGIDPTGSYHGDSDLQLERINVYYNEATGNKYVPRAILVDLEPGT 72 Query: 182 MDSVRAGPFGQLFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDIVRKEAESCDCLQG 361 MDSVR+GPFGQ+FRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLD+VRKE+ESCDCLQG Sbjct: 73 MDSVRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDVVRKESESCDCLQG 132 Query: 362 FQXXXXXXXXXXXXXXXXXXXKIREEYPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQL 541 FQ KIREEYPDRIMNTFSV+PSPKVSDTVVEPYNATLSVHQL Sbjct: 133 FQLTHSLGGGTGSGMGTLLISKIREEYPDRIMNTFSVMPSPKVSDTVVEPYNATLSVHQL 192 Query: 542 VENTDETFCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRK 721 VENTDET+CIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRK Sbjct: 193 VENTDETYCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRK 252 Query: 722 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMMAACDPRHGRYLT 901 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFD+KNMMAACDPRHGRYLT Sbjct: 253 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDSKNMMAACDPRHGRYLT 312 Query: 902 VAALFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMSATFIGNST 1081 VAA+FRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMSATFIGNST Sbjct: 313 VAAIFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMSATFIGNST 372 Query: 1082 AIQELFKRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA 1255 AIQELFKR+SEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA Sbjct: 373 AIQELFKRISEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA 430
>sp|Q7JJU6|TBB2_PANTR Tubulin beta-2 chain sp|P69895|TBB2_MACMU Tubulin beta-2 chain sp|P07437|TBB2_HUMAN Tubulin beta-2 chain sp|P09244|TBB7_CHICK Tubulin beta-7 chain (Tubulin beta 4') sp|P99024|TBB5_MOUSE Tubulin beta-5 chain sp|P69897|TBB5_RAT Tubulin beta-5 chain sp|P69893|TBB1_CRIGR Tubulin beta-1 chain (Beta-tubulin isotype I) (Class I beta tubulin) Length = 444 Score = 798 bits (2061), Expect = 0.0 Identities = 386/418 (92%), Positives = 398/418 (95%) Frame = +2 Query: 2 GNQIGAKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEATGGKYVPRAVLIDLEPGT 181 GNQIGAKFWEVISDEHGIDPTGTYHGDSDLQL+RI+VYYNEATGGKYVPRA+L+DLEPGT Sbjct: 13 GNQIGAKFWEVISDEHGIDPTGTYHGDSDLQLDRISVYYNEATGGKYVPRAILVDLEPGT 72 Query: 182 MDSVRAGPFGQLFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDIVRKEAESCDCLQG 361 MDSVR+GPFGQ+FRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLD+VRKEAESCDCLQG Sbjct: 73 MDSVRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDVVRKEAESCDCLQG 132 Query: 362 FQXXXXXXXXXXXXXXXXXXXKIREEYPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQL 541 FQ KIREEYPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQL Sbjct: 133 FQLTHSLGGGTGSGMGTLLISKIREEYPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQL 192 Query: 542 VENTDETFCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRK 721 VENTDET+CIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRK Sbjct: 193 VENTDETYCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRK 252 Query: 722 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMMAACDPRHGRYLT 901 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQ+FDAKNMMAACDPRHGRYLT Sbjct: 253 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQVFDAKNMMAACDPRHGRYLT 312 Query: 902 VAALFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMSATFIGNST 1081 VAA+FRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKM+ TFIGNST Sbjct: 313 VAAVFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMAVTFIGNST 372 Query: 1082 AIQELFKRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA 1255 AIQELFKR+SEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA Sbjct: 373 AIQELFKRISEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA 430
>sp|P09206|TBB3_CHICK Tubulin beta-3 chain (Beta-tubulin class-IV) Length = 445 Score = 797 bits (2059), Expect = 0.0 Identities = 388/418 (92%), Positives = 397/418 (94%) Frame = +2 Query: 2 GNQIGAKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEATGGKYVPRAVLIDLEPGT 181 GNQIGAKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEATGGKYVPRAVL+DLEPGT Sbjct: 13 GNQIGAKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEATGGKYVPRAVLVDLEPGT 72 Query: 182 MDSVRAGPFGQLFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDIVRKEAESCDCLQG 361 MDSVR+GPF Q+FRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLD+VRKEAESCDCLQG Sbjct: 73 MDSVRSGPFRQIFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDVVRKEAESCDCLQG 132 Query: 362 FQXXXXXXXXXXXXXXXXXXXKIREEYPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQL 541 FQ KIREEYPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQL Sbjct: 133 FQLTHSLGGGTGSGMGTLLISKIREEYPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQL 192 Query: 542 VENTDETFCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRK 721 VENTDET+CIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRK Sbjct: 193 VENTDETYCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRK 252 Query: 722 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMMAACDPRHGRYLT 901 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVP+LTQQMFDAKNMMAAC+P HGRYLT Sbjct: 253 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPDLTQQMFDAKNMMAACEPGHGRYLT 312 Query: 902 VAALFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMSATFIGNST 1081 VAA+FRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMSATFIGNST Sbjct: 313 VAAVFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMSATFIGNST 372 Query: 1082 AIQELFKRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA 1255 AIQELFKR+SEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA Sbjct: 373 AIQELFKRISEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA 430
>sp|Q91240|TBB_PSEAM Tubulin beta chain (Beta tubulin) Length = 445 Score = 797 bits (2058), Expect = 0.0 Identities = 387/418 (92%), Positives = 398/418 (95%) Frame = +2 Query: 2 GNQIGAKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEATGGKYVPRAVLIDLEPGT 181 GNQIGAKFWEVISDEHGIDP+GTYHGD++LQLERINVYYNEATGGKYVPRAVL+DLEPGT Sbjct: 13 GNQIGAKFWEVISDEHGIDPSGTYHGDNELQLERINVYYNEATGGKYVPRAVLVDLEPGT 72 Query: 182 MDSVRAGPFGQLFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDIVRKEAESCDCLQG 361 MDSVR+G FGQ+FRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLD+VRKEAESCDCLQG Sbjct: 73 MDSVRSGAFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDVVRKEAESCDCLQG 132 Query: 362 FQXXXXXXXXXXXXXXXXXXXKIREEYPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQL 541 FQ KIREEYPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQL Sbjct: 133 FQLTHSLGGGTGSGMGTLLISKIREEYPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQL 192 Query: 542 VENTDETFCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRK 721 VENTDET+CIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRK Sbjct: 193 VENTDETYCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRK 252 Query: 722 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMMAACDPRHGRYLT 901 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMMAACDPRHGRYLT Sbjct: 253 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMMAACDPRHGRYLT 312 Query: 902 VAALFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMSATFIGNST 1081 VAA+FRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMS+TFIGNST Sbjct: 313 VAAIFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMSSTFIGNST 372 Query: 1082 AIQELFKRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA 1255 AIQELFKR+SEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA Sbjct: 373 AIQELFKRMSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA 430
>sp|O17449|TBB1_MANSE Tubulin beta-1 chain (Beta-1 tubulin) Length = 447 Score = 795 bits (2054), Expect = 0.0 Identities = 383/418 (91%), Positives = 397/418 (94%) Frame = +2 Query: 2 GNQIGAKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEATGGKYVPRAVLIDLEPGT 181 GNQIGAKFWE+ISDEHGIDPTG YHGDSDLQLERINVYYNEA+GGKYVPRA+L+DLEPGT Sbjct: 13 GNQIGAKFWEIISDEHGIDPTGAYHGDSDLQLERINVYYNEASGGKYVPRAILVDLEPGT 72 Query: 182 MDSVRAGPFGQLFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDIVRKEAESCDCLQG 361 MDSVR+GPFGQ+FRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLD+VRKEAESCDCLQG Sbjct: 73 MDSVRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDVVRKEAESCDCLQG 132 Query: 362 FQXXXXXXXXXXXXXXXXXXXKIREEYPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQL 541 FQ KIREEYPDRIMNT+SVVPSPKVSDTVVEPYNATLSVHQL Sbjct: 133 FQLTHSLGGGTGSGMGTLLISKIREEYPDRIMNTYSVVPSPKVSDTVVEPYNATLSVHQL 192 Query: 542 VENTDETFCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRK 721 VENTDET+CIDNEALYDICFRTLKL+TPTYGDLNHLVS TMSGVTTCLRFPGQLNADLRK Sbjct: 193 VENTDETYCIDNEALYDICFRTLKLSTPTYGDLNHLVSLTMSGVTTCLRFPGQLNADLRK 252 Query: 722 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMMAACDPRHGRYLT 901 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMMAACDPRHGRYLT Sbjct: 253 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMMAACDPRHGRYLT 312 Query: 902 VAALFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMSATFIGNST 1081 VAA+FRGRMSMKEVDEQMLN+QNKNSSYFVEWIPNNVKTAVCDIPPRGLKMSATFIGNST Sbjct: 313 VAAIFRGRMSMKEVDEQMLNIQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMSATFIGNST 372 Query: 1082 AIQELFKRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA 1255 AIQELFKR+SEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQ+ATA Sbjct: 373 AIQELFKRISEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQEATA 430
>sp|P09203|TBB1_CHICK Tubulin beta-1 chain (Beta-tubulin class-I) Length = 445 Score = 795 bits (2054), Expect = 0.0 Identities = 384/418 (91%), Positives = 397/418 (94%) Frame = +2 Query: 2 GNQIGAKFWEVISDEHGIDPTGTYHGDSDLQLERINVYYNEATGGKYVPRAVLIDLEPGT 181 GNQIGAKFWEVISDEHGIDPTG+YHGDSDLQLERINVYYNEA G KYVPRA+L+DLEPGT Sbjct: 13 GNQIGAKFWEVISDEHGIDPTGSYHGDSDLQLERINVYYNEAAGNKYVPRAILVDLEPGT 72 Query: 182 MDSVRAGPFGQLFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDIVRKEAESCDCLQG 361 MDSVR+GPFGQ+FRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLD+VRKE+ESCDCLQG Sbjct: 73 MDSVRSGPFGQIFRPDNFVFGQSGAGNNWAKGHYTEGAELVDSVLDVVRKESESCDCLQG 132 Query: 362 FQXXXXXXXXXXXXXXXXXXXKIREEYPDRIMNTFSVVPSPKVSDTVVEPYNATLSVHQL 541 FQ KIREEYPDRIMNTFSV+PSPKVSDTVVEPYNAT+SVHQL Sbjct: 133 FQLTHSLGGGTGSGMGTLLISKIREEYPDRIMNTFSVMPSPKVSDTVVEPYNATVSVHQL 192 Query: 542 VENTDETFCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRK 721 VENTDET+CIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRK Sbjct: 193 VENTDETYCIDNEALYDICFRTLKLTTPTYGDLNHLVSATMSGVTTCLRFPGQLNADLRK 252 Query: 722 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDAKNMMAACDPRHGRYLT 901 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFD+KNMMAACDPRHGRYLT Sbjct: 253 LAVNMVPFPRLHFFMPGFAPLTSRGSQQYRALTVPELTQQMFDSKNMMAACDPRHGRYLT 312 Query: 902 VAALFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMSATFIGNST 1081 VAA+FRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMSATFIGNST Sbjct: 313 VAAIFRGRMSMKEVDEQMLNVQNKNSSYFVEWIPNNVKTAVCDIPPRGLKMSATFIGNST 372 Query: 1082 AIQELFKRVSEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA 1255 AIQELFKR+SEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA Sbjct: 373 AIQELFKRISEQFTAMFRRKAFLHWYTGEGMDEMEFTEAESNMNDLVSEYQQYQDATA 430
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 156,550,542
Number of Sequences: 369166
Number of extensions: 3168932
Number of successful extensions: 9008
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 7867
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 8289
length of database: 68,354,980
effective HSP length: 115
effective length of database: 47,110,455
effective search space used: 17477978805
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail