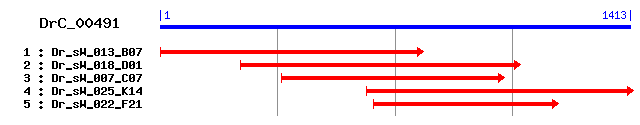
DrC_00491
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00491 (1413 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P25708|NUBM_BOVIN NADH-ubiquinone oxidoreductase 51 kDa ... 734 0.0 sp|P49821|NUBM_HUMAN NADH-ubiquinone oxidoreductase 51 kDa ... 731 0.0 sp|Q8HXQ9|NUBM_MACFA NADH-ubiquinone oxidoreductase 51 kDa ... 731 0.0 sp|Q91YT0|NUBM_MOUSE NADH-ubiquinone oxidoreductase 51 kDa ... 729 0.0 sp|P24917|NUBM_NEUCR NADH-ubiquinone oxidoreductase 51 kDa ... 665 0.0 sp|Q92406|NUBM_ASPNG NADH-ubiquinone oxidoreductase 51 kDa ... 637 0.0 sp|P56912|NUOF1_RHIME NADH-quinone oxidoreductase chain F 1... 606 e-173 sp|Q9ZE33|NUOF_RICPR NADH-quinone oxidoreductase chain F (N... 601 e-171 sp|O07948|NUOF_RHOCA NADH-quinone oxidoreductase chain F (N... 593 e-169 sp|P29913|NQO1_PARDE NADH-quinone oxidoreductase chain 1 (N... 583 e-166
>sp|P25708|NUBM_BOVIN NADH-ubiquinone oxidoreductase 51 kDa subunit, mitochondrial precursor (Complex I-51KD) (CI-51KD) (NADH dehydrogenase flavoprotein 1) Length = 464 Score = 734 bits (1896), Expect = 0.0 Identities = 340/419 (81%), Positives = 381/419 (90%), Gaps = 1/419 (0%) Frame = +3 Query: 120 LKDEDRIFSNLYGRYDWHLQKAMSRGHWFKTKEIILKGHEWILNEIKISGLRGRGGAGFP 299 LKDEDRIF+NLYGR+DW L+ A SRG W+KTKEI+LKG +WIL E+K SGLRGRGGAGFP Sbjct: 35 LKDEDRIFTNLYGRHDWRLKGAQSRGDWYKTKEILLKGPDWILGEVKTSGLRGRGGAGFP 94 Query: 300 TGMKWGFMNKPSD-RPKYLVVNADEGEPGTCKDREIMRHDPHTLVEGCLIAGKAMGAVAA 476 TG+KW FMNKPSD RPKYLVVNADEGEPGTCKDREI+RHDPH LVEGCL+ G+AMGA AA Sbjct: 95 TGLKWSFMNKPSDGRPKYLVVNADEGEPGTCKDREIIRHDPHKLVEGCLVGGRAMGARAA 154 Query: 477 YIYIRGEFYNEASNMQVAINEAYKAGFIGKNACGTGFDFDIFMHRGAGAYICGEETALIE 656 YIYIRGEFYNEASN+QVAI EAY+AG IGKNACG+G+DFD+F+ RGAGAYICGEETALIE Sbjct: 155 YIYIRGEFYNEASNLQVAIREAYEAGLIGKNACGSGYDFDVFVVRGAGAYICGEETALIE 214 Query: 657 SIEGKQGKPRLKPPFPADIGLFGCPTTVSNVETVAVAPTICRRGGEWFAQFGRERNKGTK 836 SIEGKQGKPRLKPPFPAD+G+FGCPTTV+NVETVAV+PTICRRGG WFA FGRERN GTK Sbjct: 215 SIEGKQGKPRLKPPFPADVGVFGCPTTVANVETVAVSPTICRRGGAWFASFGRERNSGTK 274 Query: 837 LFNISGHVNNPCTVEETMSIPLKALIEKHCHGIIGGWDNLLAVIPGGSSTPLIPKNVCDS 1016 LFNISGHVNNPCTVEE MS+PLK LIEKH G+ GGWDNLLAVIPGGSSTPLIPK+VC++ Sbjct: 275 LFNISGHVNNPCTVEEEMSVPLKELIEKHAGGVTGGWDNLLAVIPGGSSTPLIPKSVCET 334 Query: 1017 VLMDFDSLVEAQTALGTAALIVMNKDADVVRCIARLMDFYKHESCGQCTPCREGTGWMQK 1196 VLMDFD+L++AQT LGTAA+IVM++ D+V+ IARL++FYKHESCGQCTPCREG WM K Sbjct: 335 VLMDFDALIQAQTGLGTAAVIVMDRSTDIVKAIARLIEFYKHESCGQCTPCREGVDWMNK 394 Query: 1197 IVKRFVNGNAVKPEIDMLWEISKQIEGHTICALGDGAAWPVQGLIRHFRPELEMRMKNY 1373 ++ RFV G+A EID LWEISKQIEGHTICALGDGAAWPVQGLIRHFRPELE RM+ + Sbjct: 395 VMARFVRGDARPAEIDSLWEISKQIEGHTICALGDGAAWPVQGLIRHFRPELEERMQQF 453
>sp|P49821|NUBM_HUMAN NADH-ubiquinone oxidoreductase 51 kDa subunit, mitochondrial precursor (Complex I-51KD) (CI-51KD) (NADH dehydrogenase flavoprotein 1) Length = 464 Score = 731 bits (1888), Expect = 0.0 Identities = 339/419 (80%), Positives = 382/419 (91%), Gaps = 1/419 (0%) Frame = +3 Query: 120 LKDEDRIFSNLYGRYDWHLQKAMSRGHWFKTKEIILKGHEWILNEIKISGLRGRGGAGFP 299 LKDEDRIF+NLYGR+DW L+ ++SRG W+KTKEI+LKG +WIL EIK SGLRGRGGAGFP Sbjct: 35 LKDEDRIFTNLYGRHDWRLKGSLSRGDWYKTKEILLKGPDWILGEIKTSGLRGRGGAGFP 94 Query: 300 TGMKWGFMNKPSD-RPKYLVVNADEGEPGTCKDREIMRHDPHTLVEGCLIAGKAMGAVAA 476 TG+KW FMNKPSD RPKYLVVNADEGEPGTCKDREI+RHDPH L+EGCL+ G+AMGA AA Sbjct: 95 TGLKWSFMNKPSDGRPKYLVVNADEGEPGTCKDREILRHDPHKLLEGCLVGGRAMGARAA 154 Query: 477 YIYIRGEFYNEASNMQVAINEAYKAGFIGKNACGTGFDFDIFMHRGAGAYICGEETALIE 656 YIYIRGEFYNEASN+QVAI EAY+AG IGKNACG+G+DFD+F+ RGAGAYICGEETALIE Sbjct: 155 YIYIRGEFYNEASNLQVAIREAYEAGLIGKNACGSGYDFDVFVVRGAGAYICGEETALIE 214 Query: 657 SIEGKQGKPRLKPPFPADIGLFGCPTTVSNVETVAVAPTICRRGGEWFAQFGRERNKGTK 836 SIEGKQGKPRLKPPFPAD+G+FGCPTTV+NVETVAV+PTICRRGG WFA FGRERN GTK Sbjct: 215 SIEGKQGKPRLKPPFPADVGVFGCPTTVANVETVAVSPTICRRGGTWFAGFGRERNSGTK 274 Query: 837 LFNISGHVNNPCTVEETMSIPLKALIEKHCHGIIGGWDNLLAVIPGGSSTPLIPKNVCDS 1016 LFNISGHVN+PCTVEE MS+PLK LIEKH G+ GGWDNLLAVIPGGSSTPLIPK+VC++ Sbjct: 275 LFNISGHVNHPCTVEEEMSVPLKELIEKHAGGVTGGWDNLLAVIPGGSSTPLIPKSVCET 334 Query: 1017 VLMDFDSLVEAQTALGTAALIVMNKDADVVRCIARLMDFYKHESCGQCTPCREGTGWMQK 1196 VLMDFD+LV+AQT LGTAA+IVM++ D+V+ IARL++FYKHESCGQCTPCREG WM K Sbjct: 335 VLMDFDALVQAQTGLGTAAVIVMDRSTDIVKAIARLIEFYKHESCGQCTPCREGVDWMNK 394 Query: 1197 IVKRFVNGNAVKPEIDMLWEISKQIEGHTICALGDGAAWPVQGLIRHFRPELEMRMKNY 1373 ++ RFV G+A EID LWEISKQIEGHTICALGDGAAWPVQGLIRHFRPELE RM+ + Sbjct: 395 VMARFVRGDARPAEIDSLWEISKQIEGHTICALGDGAAWPVQGLIRHFRPELEERMQRF 453
>sp|Q8HXQ9|NUBM_MACFA NADH-ubiquinone oxidoreductase 51 kDa subunit, mitochondrial precursor (Complex I-51KD) (CI-51KD) (NADH dehydrogenase flavoprotein 1) Length = 464 Score = 731 bits (1887), Expect = 0.0 Identities = 340/419 (81%), Positives = 382/419 (91%), Gaps = 1/419 (0%) Frame = +3 Query: 120 LKDEDRIFSNLYGRYDWHLQKAMSRGHWFKTKEIILKGHEWILNEIKISGLRGRGGAGFP 299 LKDEDRIF+NLYGR+DW L+ A+SRG W+KTKEI+LKG +WIL EIK SGLRGRGGAGFP Sbjct: 35 LKDEDRIFTNLYGRHDWRLKGALSRGDWYKTKEILLKGPDWILGEIKTSGLRGRGGAGFP 94 Query: 300 TGMKWGFMNKPSD-RPKYLVVNADEGEPGTCKDREIMRHDPHTLVEGCLIAGKAMGAVAA 476 TG+KW FMNKPSD RPKYLVVNADEGEPGTCKDREI+RHDPH LVEGCL+ G+AMGA AA Sbjct: 95 TGLKWSFMNKPSDGRPKYLVVNADEGEPGTCKDREIIRHDPHKLVEGCLVGGRAMGARAA 154 Query: 477 YIYIRGEFYNEASNMQVAINEAYKAGFIGKNACGTGFDFDIFMHRGAGAYICGEETALIE 656 YIYIRGEFYNEASN+QVAI EAY+AG IGKNACG+G+DFD+F+ RGAGAYICGEETALIE Sbjct: 155 YIYIRGEFYNEASNLQVAIREAYEAGLIGKNACGSGYDFDVFVVRGAGAYICGEETALIE 214 Query: 657 SIEGKQGKPRLKPPFPADIGLFGCPTTVSNVETVAVAPTICRRGGEWFAQFGRERNKGTK 836 SIEGKQGKPRLKPPFPAD+G+FGCPTTV+NVETVAV+PTICRRGG WFA FGRERN GTK Sbjct: 215 SIEGKQGKPRLKPPFPADVGVFGCPTTVANVETVAVSPTICRRGGTWFAGFGRERNSGTK 274 Query: 837 LFNISGHVNNPCTVEETMSIPLKALIEKHCHGIIGGWDNLLAVIPGGSSTPLIPKNVCDS 1016 LFNISGHVN+PCTVEE MS+PLK LIEKH G+ GGWDNLLAVIPGGSSTPLIPK+VC++ Sbjct: 275 LFNISGHVNHPCTVEEEMSVPLKELIEKHAGGVTGGWDNLLAVIPGGSSTPLIPKSVCET 334 Query: 1017 VLMDFDSLVEAQTALGTAALIVMNKDADVVRCIARLMDFYKHESCGQCTPCREGTGWMQK 1196 VLMDFD+LV+AQT LGTAA+IVM++ D+V+ IARL++FYK+ESCGQCTPCREG WM K Sbjct: 335 VLMDFDALVQAQTGLGTAAVIVMDRSTDIVKAIARLIEFYKYESCGQCTPCREGVDWMNK 394 Query: 1197 IVKRFVNGNAVKPEIDMLWEISKQIEGHTICALGDGAAWPVQGLIRHFRPELEMRMKNY 1373 ++ RFV G+A EID LWEISKQIEGHTICALGDGAAWPVQGLIRHFRPELE RM+ + Sbjct: 395 VMARFVRGDARPAEIDSLWEISKQIEGHTICALGDGAAWPVQGLIRHFRPELEERMQRF 453
>sp|Q91YT0|NUBM_MOUSE NADH-ubiquinone oxidoreductase 51 kDa subunit, mitochondrial precursor (Complex I-51KD) (CI-51KD) (NADH dehydrogenase flavoprotein 1) Length = 464 Score = 729 bits (1881), Expect = 0.0 Identities = 339/419 (80%), Positives = 380/419 (90%), Gaps = 1/419 (0%) Frame = +3 Query: 120 LKDEDRIFSNLYGRYDWHLQKAMSRGHWFKTKEIILKGHEWILNEIKISGLRGRGGAGFP 299 LKDEDRIF+NLYGR+DW L+ A+ RG W+KTKEI+LKG +WIL E+K SGLRGRGGAGFP Sbjct: 35 LKDEDRIFTNLYGRHDWRLKGALRRGDWYKTKEILLKGPDWILGEMKTSGLRGRGGAGFP 94 Query: 300 TGMKWGFMNKPSD-RPKYLVVNADEGEPGTCKDREIMRHDPHTLVEGCLIAGKAMGAVAA 476 TG+KW FMNKPSD RPKYLVVNADEGEPGTCKDREIMRHDPH LVEGCL+ G+AMGA AA Sbjct: 95 TGLKWSFMNKPSDGRPKYLVVNADEGEPGTCKDREIMRHDPHKLVEGCLVGGRAMGARAA 154 Query: 477 YIYIRGEFYNEASNMQVAINEAYKAGFIGKNACGTGFDFDIFMHRGAGAYICGEETALIE 656 YIYIRGEFYNEASN+QVAI EAY+AG IGKNACG+ +DFD+F+ RGAGAYICGEETALIE Sbjct: 155 YIYIRGEFYNEASNLQVAIREAYEAGLIGKNACGSDYDFDVFVVRGAGAYICGEETALIE 214 Query: 657 SIEGKQGKPRLKPPFPADIGLFGCPTTVSNVETVAVAPTICRRGGEWFAQFGRERNKGTK 836 SIEGKQGKPRLKPPFPAD+G+FGCPTTV+NVETVAV+PTICRRGG WFA FGRERN GTK Sbjct: 215 SIEGKQGKPRLKPPFPADVGVFGCPTTVANVETVAVSPTICRRGGTWFAGFGRERNSGTK 274 Query: 837 LFNISGHVNNPCTVEETMSIPLKALIEKHCHGIIGGWDNLLAVIPGGSSTPLIPKNVCDS 1016 LFNISGHVN+PCTVEE MS+PLK LIEKH G+ GGWDNLLAVIPGGSSTPLIPK+VC++ Sbjct: 275 LFNISGHVNHPCTVEEEMSVPLKELIEKHAGGVTGGWDNLLAVIPGGSSTPLIPKSVCET 334 Query: 1017 VLMDFDSLVEAQTALGTAALIVMNKDADVVRCIARLMDFYKHESCGQCTPCREGTGWMQK 1196 VLMDFD+LV+AQT LGTAA+IVM++ D+V+ IARL++FYKHESCGQCTPCREG WM K Sbjct: 335 VLMDFDALVQAQTGLGTAAVIVMDRSTDIVKAIARLIEFYKHESCGQCTPCREGVDWMNK 394 Query: 1197 IVKRFVNGNAVKPEIDMLWEISKQIEGHTICALGDGAAWPVQGLIRHFRPELEMRMKNY 1373 ++ RFV G+A EID LWEISKQIEGHTICALGDGAAWPVQGLIRHFRPELE RM+ + Sbjct: 395 VMARFVKGDARPAEIDSLWEISKQIEGHTICALGDGAAWPVQGLIRHFRPELEDRMQRF 453
>sp|P24917|NUBM_NEUCR NADH-ubiquinone oxidoreductase 51 kDa subunit, mitochondrial precursor (Complex I-51KD) (CI-51KD) Length = 493 Score = 665 bits (1717), Expect = 0.0 Identities = 306/422 (72%), Positives = 361/422 (85%), Gaps = 4/422 (0%) Frame = +3 Query: 120 LKDEDRIFSNLYGRYDWHLQKAMSRGHWFKTKEIILKGHEWILNEIKISGLRGRGGAGFP 299 LKD+DRIF NLYGRY L+ A G W KTKEI+LKGH+WI+ E+K SGLRGRGGAGFP Sbjct: 44 LKDQDRIFQNLYGRYPPDLKHAKKMGDWHKTKEILLKGHDWIIGEVKASGLRGRGGAGFP 103 Query: 300 TGMKWGFMN----KPSDRPKYLVVNADEGEPGTCKDREIMRHDPHTLVEGCLIAGKAMGA 467 +G+KW FMN D+P+YLVVNADEGEPGTCKDREIMR DPH LVEGCL+AG+AM A Sbjct: 104 SGLKWSFMNFKDWDKDDKPRYLVVNADEGEPGTCKDREIMRKDPHKLVEGCLVAGRAMNA 163 Query: 468 VAAYIYIRGEFYNEASNMQVAINEAYKAGFIGKNACGTGFDFDIFMHRGAGAYICGEETA 647 AAYIYIRGEF EA+ +Q AINEAY G IGKNACG+G+DFD+++HRGAGAY+CGEET+ Sbjct: 164 TAAYIYIRGEFIQEAAILQNAINEAYADGLIGKNACGSGYDFDVYLHRGAGAYVCGEETS 223 Query: 648 LIESIEGKQGKPRLKPPFPADIGLFGCPTTVSNVETVAVAPTICRRGGEWFAQFGRERNK 827 LIES+EGK GKPRLKPPFPA +GLFGCP+TV+NVETVAVAPTICRRGG WFA FGRERN+ Sbjct: 224 LIESLEGKPGKPRLKPPFPAAVGLFGCPSTVANVETVAVAPTICRRGGNWFAGFGRERNQ 283 Query: 828 GTKLFNISGHVNNPCTVEETMSIPLKALIEKHCHGIIGGWDNLLAVIPGGSSTPLIPKNV 1007 GTKLF ISGHVNNPCTVEE MSIP++ LI+KHC G+ GGWDNLLAVIPGGSSTP++PK++ Sbjct: 284 GTKLFCISGHVNNPCTVEEEMSIPMRELIDKHCGGVRGGWDNLLAVIPGGSSTPILPKHI 343 Query: 1008 CDSVLMDFDSLVEAQTALGTAALIVMNKDADVVRCIARLMDFYKHESCGQCTPCREGTGW 1187 CD+ LMDFD+L ++Q+ LGTAALIVM+K DVVR I+RL FY+HESCGQCTPCREG+ W Sbjct: 344 CDTQLMDFDALKDSQSGLGTAALIVMDKSTDVVRAISRLSHFYRHESCGQCTPCREGSKW 403 Query: 1188 MQKIVKRFVNGNAVKPEIDMLWEISKQIEGHTICALGDGAAWPVQGLIRHFRPELEMRMK 1367 ++I+KRF G + EIDML E++KQ+EGHTICALG+ AWP+QGLIRHFRPELE R++ Sbjct: 404 TEQIMKRFEKGQGREREIDMLQELTKQVEGHTICALGEAFAWPIQGLIRHFRPELEARIR 463 Query: 1368 NY 1373 + Sbjct: 464 KF 465
>sp|Q92406|NUBM_ASPNG NADH-ubiquinone oxidoreductase 51 kDa subunit, mitochondrial precursor (Complex I-51KD) (CI-51KD) Length = 496 Score = 637 bits (1643), Expect = 0.0 Identities = 292/422 (69%), Positives = 353/422 (83%), Gaps = 4/422 (0%) Frame = +3 Query: 120 LKDEDRIFSNLYGRYDWHLQKAMSRGHWFKTKEIILKGHEWILNEIKISGLRGRGGAGFP 299 LKD+DRIF+NLYG + L+ AM G W +TK+I+LKGH+W+++E+K SGLRGRGGAGFP Sbjct: 46 LKDQDRIFTNLYGHHGADLKSAMKYGDWHRTKDIVLKGHDWLISELKASGLRGRGGAGFP 105 Query: 300 TGMKWGFMN----KPSDRPKYLVVNADEGEPGTCKDREIMRHDPHTLVEGCLIAGKAMGA 467 +G+K+ FMN RP+YLVVNADEGEPGTCKDREIMR DP L+EGCL+ G+AM A Sbjct: 106 SGLKYSFMNFKDWDKDPRPRYLVVNADEGEPGTCKDREIMRKDPQKLIEGCLVVGRAMNA 165 Query: 468 VAAYIYIRGEFYNEASNMQVAINEAYKAGFIGKNACGTGFDFDIFMHRGAGAYICGEETA 647 AAY+YIRGEFY EA+ +Q AINEAY+AG IGKNACGTG+DFD+++HRG GAY+CGEET+ Sbjct: 166 NAAYMYIRGEFYQEATVLQRAINEAYEAGLIGKNACGTGYDFDVYIHRGMGAYVCGEETS 225 Query: 648 LIESIEGKQGKPRLKPPFPADIGLFGCPTTVSNVETVAVAPTICRRGGEWFAQFGRERNK 827 LIESIEGK GKPRLKPPFPA +GLFGCP+TV+NVETVAV PTI RRG WF+ FGRERN Sbjct: 226 LIESIEGKAGKPRLKPPFPAAVGLFGCPSTVTNVETVAVTPTIMRRGASWFSSFGRERNA 285 Query: 828 GTKLFNISGHVNNPCTVEETMSIPLKALIEKHCHGIIGGWDNLLAVIPGGSSTPLIPKNV 1007 GTKLF ISGHVNNPCTVEE MSI L+ +I++HC G+ GGWDNLLAVIPGGSSTP++PK + Sbjct: 286 GTKLFCISGHVNNPCTVEEEMSISLRDVIDRHCGGVRGGWDNLLAVIPGGSSTPVLPKTI 345 Query: 1008 CDSVLMDFDSLVEAQTALGTAALIVMNKDADVVRCIARLMDFYKHESCGQCTPCREGTGW 1187 CD LMDFD+L ++Q+ LGTAA+IVM+K D+VR I+RL FYKHESCGQCTPCREG+ W Sbjct: 346 CDDQLMDFDALKDSQSGLGTAAVIVMDKSTDIVRAISRLSTFYKHESCGQCTPCREGSKW 405 Query: 1188 MQKIVKRFVNGNAVKPEIDMLWEISKQIEGHTICALGDGAAWPVQGLIRHFRPELEMRMK 1367 +++R G A + EIDML E++KQ+EGHTICALG+ AWP+QGLIRHFRPELE R++ Sbjct: 406 TMHMMQRMEKGQAREREIDMLQELTKQVEGHTICALGEAFAWPIQGLIRHFRPELEARIR 465 Query: 1368 NY 1373 Y Sbjct: 466 EY 467
>sp|P56912|NUOF1_RHIME NADH-quinone oxidoreductase chain F 1 (NADH dehydrogenase I, chain F 1) (NDH-1, chain F 1) Length = 434 Score = 606 bits (1563), Expect = e-173 Identities = 279/419 (66%), Positives = 338/419 (80%), Gaps = 1/419 (0%) Frame = +3 Query: 120 LKDEDRIFSNLYGRYDWHLQKAMSRGHWFKTKEIILKGHEWILNEIKISGLRGRGGAGFP 299 L+DEDRIF+N+YG D L+ AM+RGHW TK+ + KG +WI+NE+K SGLRGRGGAGFP Sbjct: 2 LRDEDRIFTNIYGLKDKSLRGAMARGHWDGTKQFLEKGRDWIINEVKASGLRGRGGAGFP 61 Query: 300 TGMKWGFMNKPSD-RPKYLVVNADEGEPGTCKDREIMRHDPHTLVEGCLIAGKAMGAVAA 476 TG+KW FM K SD RP YLVVNADE EPGTCKDR+IMRHDPHTL+EGC+IA AMGA AA Sbjct: 62 TGLKWSFMPKESDGRPHYLVVNADESEPGTCKDRDIMRHDPHTLIEGCVIASFAMGAHAA 121 Query: 477 YIYIRGEFYNEASNMQVAINEAYKAGFIGKNACGTGFDFDIFMHRGAGAYICGEETALIE 656 YIY+RGEF E +Q AI+E Y+ G +GKN G+D DI++H GAGAYICGEETAL+E Sbjct: 122 YIYVRGEFIREREALQAAIDECYEYGLLGKNN-KLGYDIDIYVHHGAGAYICGEETALLE 180 Query: 657 SIEGKQGKPRLKPPFPADIGLFGCPTTVSNVETVAVAPTICRRGGEWFAQFGRERNKGTK 836 S+EGK+G+PRLKPPFPA++GL+GCPTTV+NVE++AV PTI RRG W+ FGR N GTK Sbjct: 181 SLEGKKGQPRLKPPFPANMGLYGCPTTVNNVESIAVTPTILRRGAGWYTSFGRPNNHGTK 240 Query: 837 LFNISGHVNNPCTVEETMSIPLKALIEKHCHGIIGGWDNLLAVIPGGSSTPLIPKNVCDS 1016 L+++SGHVN PCTVE+ MSIP LIEKHC GI GGWDNLLAVIPGGSS P +P Sbjct: 241 LYSVSGHVNRPCTVEDAMSIPFHELIEKHCGGIRGGWDNLLAVIPGGSSVPCVPGAQMKD 300 Query: 1017 VLMDFDSLVEAQTALGTAALIVMNKDADVVRCIARLMDFYKHESCGQCTPCREGTGWMQK 1196 +MD+D L E + LGTAA+IVM+K D+++ I RL FYKHESCGQCTPCREGTGWM + Sbjct: 301 AIMDYDGLRELGSGLGTAAVIVMDKSTDIIKAIWRLSAFYKHESCGQCTPCREGTGWMMR 360 Query: 1197 IVKRFVNGNAVKPEIDMLWEISKQIEGHTICALGDGAAWPVQGLIRHFRPELEMRMKNY 1373 +++R V G A K EIDML++++KQ+EGHTICALGD AAWP+QGLI+HFRPE+E R+ Y Sbjct: 361 VMERMVQGRAQKREIDMLFDVTKQVEGHTICALGDAAAWPIQGLIKHFRPEMEKRIDEY 419
>sp|Q9ZE33|NUOF_RICPR NADH-quinone oxidoreductase chain F (NADH dehydrogenase I, chain F) (NDH-1, chain F) Length = 421 Score = 601 bits (1550), Expect = e-171 Identities = 275/418 (65%), Positives = 339/418 (81%) Frame = +3 Query: 120 LKDEDRIFSNLYGRYDWHLQKAMSRGHWFKTKEIILKGHEWILNEIKISGLRGRGGAGFP 299 LK+ED+IF+NLYG+ + L+ + RG W+ TK +I KG ++I++E+K SGLRGRGGAGF Sbjct: 2 LKEEDKIFTNLYGQQSYDLKSSQKRGDWYNTKALIDKGRDFIIDEVKKSGLRGRGGAGFS 61 Query: 300 TGMKWGFMNKPSDRPKYLVVNADEGEPGTCKDREIMRHDPHTLVEGCLIAGKAMGAVAAY 479 TGMKW FM K S++P YLVVNADE EPGTCKDR+I+R +PH L+EGCLIA A+GA Y Sbjct: 62 TGMKWSFMPKNSEKPCYLVVNADESEPGTCKDRDILRFEPHKLIEGCLIASFAIGANTCY 121 Query: 480 IYIRGEFYNEASNMQVAINEAYKAGFIGKNACGTGFDFDIFMHRGAGAYICGEETALIES 659 IYIRGEFYNEASNMQ A++EAYK G IGKNACG+GFD +I++HRGAGAYICGEETAL+ES Sbjct: 122 IYIRGEFYNEASNMQRALDEAYKEGLIGKNACGSGFDCNIYLHRGAGAYICGEETALLES 181 Query: 660 IEGKQGKPRLKPPFPADIGLFGCPTTVSNVETVAVAPTICRRGGEWFAQFGRERNKGTKL 839 +EGK+G PRLKPPFPA GL+GCPTT++NVE++AV PTI RRG WFA G+ N GTK+ Sbjct: 182 LEGKKGMPRLKPPFPAGFGLYGCPTTINNVESIAVVPTILRRGASWFAAIGKPNNTGTKV 241 Query: 840 FNISGHVNNPCTVEETMSIPLKALIEKHCHGIIGGWDNLLAVIPGGSSTPLIPKNVCDSV 1019 F ISGHVN PC +EE M +PLK LIEK+ G+ GGW+NL A+IPGG+S PL+PK++C+ V Sbjct: 242 FCISGHVNKPCNIEEVMGVPLKELIEKYAGGVRGGWNNLKAIIPGGASVPLLPKSLCE-V 300 Query: 1020 LMDFDSLVEAQTALGTAALIVMNKDADVVRCIARLMDFYKHESCGQCTPCREGTGWMQKI 1199 MDFDSL + LGT +IVM+K D++ IARL FY HESCGQCTPCREGTGWM ++ Sbjct: 301 EMDFDSLRTVGSGLGTGGIIVMDKSTDIIYAIARLSKFYMHESCGQCTPCREGTGWMWRV 360 Query: 1200 VKRFVNGNAVKPEIDMLWEISKQIEGHTICALGDGAAWPVQGLIRHFRPELEMRMKNY 1373 + R VNGNA K EID L ++K+IEGHTICALGD AAWP+QGLIRHFR E+E R+K++ Sbjct: 361 MMRLVNGNAKKNEIDTLLNVTKEIEGHTICALGDAAAWPIQGLIRHFRDEIEQRIKSF 418
>sp|O07948|NUOF_RHOCA NADH-quinone oxidoreductase chain F (NADH dehydrogenase I, chain F) (NDH-1, chain F) Length = 431 Score = 593 bits (1530), Expect = e-169 Identities = 272/420 (64%), Positives = 334/420 (79%), Gaps = 1/420 (0%) Frame = +3 Query: 120 LKDEDRIFSNLYGRYDWHLQKAMSRGHWFKTKEIILKGHEWILNEIKISGLRGRGGAGFP 299 L D DRIF+N+YG +D L+ AM+RGHW T I+ G +WI+ ++K SGLRGRGGAGFP Sbjct: 2 LNDHDRIFTNIYGMHDRSLKGAMARGHWDGTAAILGNGRDWIIEQVKASGLRGRGGAGFP 61 Query: 300 TGMKWGFMNKPSD-RPKYLVVNADEGEPGTCKDREIMRHDPHTLVEGCLIAGKAMGAVAA 476 TG+KW FM K SD RP +LV+NADE EPGTCKDREIMRHDPHTL+EG LIAG AMGA AA Sbjct: 62 TGLKWSFMPKQSDGRPSFLVINADESEPGTCKDREIMRHDPHTLIEGALIAGFAMGARAA 121 Query: 477 YIYIRGEFYNEASNMQVAINEAYKAGFIGKNACGTGFDFDIFMHRGAGAYICGEETALIE 656 YIYIRGE+ E +Q AI+E Y+AG IGKNAC + +DFD+++H GAGAYICGEETAL+E Sbjct: 122 YIYIRGEYVREKEALQAAIDECYEAGLIGKNACRSDYDFDVYLHHGAGAYICGEETALLE 181 Query: 657 SIEGKQGKPRLKPPFPADIGLFGCPTTVSNVETVAVAPTICRRGGEWFAQFGRERNKGTK 836 S+EGK+G PR+KPPFPA GL+GCPTTV+NVE++AV PTI RRGG WFA GR N G K Sbjct: 182 SLEGKKGMPRMKPPFPAGSGLYGCPTTVNNVESIAVVPTILRRGGAWFAGIGRPNNTGVK 241 Query: 837 LFNISGHVNNPCTVEETMSIPLKALIEKHCHGIIGGWDNLLAVIPGGSSTPLIPKNVCDS 1016 LF +SGHVN PC VEE MSI +K LIEKH G+ GGW NL AVIPGG+S P+IP ++C+ Sbjct: 242 LFAMSGHVNTPCVVEEAMSISMKELIEKHGGGVRGGWKNLKAVIPGGASCPVIPAHLCED 301 Query: 1017 VLMDFDSLVEAQTALGTAALIVMNKDADVVRCIARLMDFYKHESCGQCTPCREGTGWMQK 1196 +MD+D + E Q++ GTA LIVM++ D+++ IARL F+KHESCGQCTPCREGT WM + Sbjct: 302 AIMDYDGMREKQSSFGTACLIVMDQQTDIIKAIARLSKFFKHESCGQCTPCREGTSWMWR 361 Query: 1197 IVKRFVNGNAVKPEIDMLWEISKQIEGHTICALGDGAAWPVQGLIRHFRPELEMRMKNYK 1376 +++R V G A EIDML+ ++KQ+EGHTICALGD AAWP+QGLIRH+R E+E R+K K Sbjct: 362 VMERLVTGEAEVEEIDMLFSVTKQVEGHTICALGDAAAWPIQGLIRHYREEIEDRIKARK 421
>sp|P29913|NQO1_PARDE NADH-quinone oxidoreductase chain 1 (NADH dehydrogenase I, chain 1) (NDH-1, chain 1) Length = 431 Score = 583 bits (1503), Expect = e-166 Identities = 270/417 (64%), Positives = 331/417 (79%), Gaps = 1/417 (0%) Frame = +3 Query: 120 LKDEDRIFSNLYGRYDWHLQKAMSRGHWFKTKEIILKGHEWILNEIKISGLRGRGGAGFP 299 L D+DRIF+NLYG D L A RGHW T II +G + I++E+K SGLRGRGGAGFP Sbjct: 2 LNDQDRIFTNLYGMGDRSLAGAKKRGHWDGTAAIIQRGRDKIIDEMKASGLRGRGGAGFP 61 Query: 300 TGMKWGFMNKPSD-RPKYLVVNADEGEPGTCKDREIMRHDPHTLVEGCLIAGKAMGAVAA 476 TGMKW FM K SD RP YLV+NADE EP TCKDREIMRHDPHTL+EG LIA AMGA AA Sbjct: 62 TGMKWSFMPKESDGRPSYLVINADESEPATCKDREIMRHDPHTLIEGALIASFAMGAHAA 121 Query: 477 YIYIRGEFYNEASNMQVAINEAYKAGFIGKNACGTGFDFDIFMHRGAGAYICGEETALIE 656 YIYIRGEF E +Q AI+E Y AG +G+NA G+G+DFD+++H GAGAYICGEETAL+E Sbjct: 122 YIYIRGEFIREREALQAAIDECYDAGLLGRNAAGSGWDFDLYLHHGAGAYICGEETALLE 181 Query: 657 SIEGKQGKPRLKPPFPADIGLFGCPTTVSNVETVAVAPTICRRGGEWFAQFGRERNKGTK 836 S+EGK+G PR+KPPFPA GL+GCPTTV+NVE++AV PTI RRG EWFA FGR N G K Sbjct: 182 SLEGKKGMPRMKPPFPAGAGLYGCPTTVNNVESIAVVPTILRRGAEWFASFGRPNNAGVK 241 Query: 837 LFNISGHVNNPCTVEETMSIPLKALIEKHCHGIIGGWDNLLAVIPGGSSTPLIPKNVCDS 1016 LF ++GHVN PC VEE MSIP++ LIEKH GI GGW NL AVIPGG+S P++ C++ Sbjct: 242 LFGLTGHVNTPCVVEEAMSIPMRELIEKHGGGIRGGWKNLKAVIPGGASCPVLTAEQCEN 301 Query: 1017 VLMDFDSLVEAQTALGTAALIVMNKDADVVRCIARLMDFYKHESCGQCTPCREGTGWMQK 1196 +MD+D + + +++ GTA +IVM++ DVV+ I RL F+KHESCGQCTPCREGTGWM + Sbjct: 302 AIMDYDGMRDVRSSFGTACMIVMDQSTDVVKAIWRLSKFFKHESCGQCTPCREGTGWMMR 361 Query: 1197 IVKRFVNGNAVKPEIDMLWEISKQIEGHTICALGDGAAWPVQGLIRHFRPELEMRMK 1367 +++R V G+A EIDML++++KQ+EGHTICALGD AAWP+QGLIR+FR E+E R+K Sbjct: 362 VMERLVRGDAEVEEIDMLFDVTKQVEGHTICALGDAAAWPIQGLIRNFREEIEDRIK 418
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 166,886,371
Number of Sequences: 369166
Number of extensions: 3725299
Number of successful extensions: 9639
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 9040
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 9580
length of database: 68,354,980
effective HSP length: 114
effective length of database: 47,295,190
effective search space used: 16837087640
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail