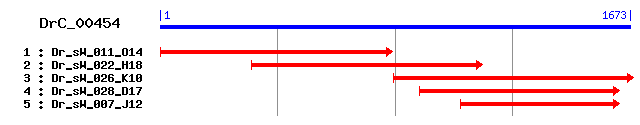
DrC_00454
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00454 (1672 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q01406|SRC8_CHICK Src substrate protein p85 (p80) (Corta... 458 e-128 sp|Q60598|SRC8_MOUSE Src substrate cortactin 451 e-126 sp|Q14247|SRC8_HUMAN Src substrate cortactin (Amplaxin) (On... 449 e-125 sp|P14317|HCLS1_HUMAN Hematopoietic lineage cell-specific p... 290 1e-77 sp|P49710|HCLS1_MOUSE Hematopoietic lineage cell-specific p... 287 7e-77 sp|Q6XPR3|RPTN_HUMAN Repetin 67 2e-10 sp|Q925Q9|SH3K1_RAT SH3-domain kinase binding protein 1 (SH... 67 2e-10 sp|Q8R550|SH3K1_MOUSE SH3-domain kinase binding protein 1 (... 66 3e-10 sp|Q14847|LASP1_HUMAN LIM and SH3 domain protein 1 (LASP-1)... 66 3e-10 sp|Q99MZ8|LASP1_RAT LIM and SH3 domain protein 1 (LASP-1) 65 4e-10
>sp|Q01406|SRC8_CHICK Src substrate protein p85 (p80) (Cortactin) Length = 563 Score = 458 bits (1179), Expect = e-128 Identities = 250/558 (44%), Positives = 337/558 (60%), Gaps = 59/558 (10%) Frame = +3 Query: 36 INMWKATAGSXXXXXXXXXXXXXXXXXXXXFQNDVTEKEQRWGSKTVSGSGRTDFIDLSA 215 + MWKATAG F NDV+EKEQRWG+KTV GSG + I++ Sbjct: 9 LQMWKATAGHSIAVSQDDGADDWETDPD--FVNDVSEKEQRWGAKTVKGSGHQEHINIHQ 66 Query: 216 LKESVKKSDADNKKIAYDRSSKPSYGYGGSFGVQKDRMDKCAVGNDYVAAVDKHTSQKDY 395 L+E+V + K+ + K S+GYGG FGV++DRMDK AVG++Y + + KH SQ D Sbjct: 67 LRENVFQEHQTIKEKELETGPKASHGYGGKFGVEQDRMDKSAVGHEYQSKLSKHCSQVDS 126 Query: 396 SQGFGGKYGVQSDRKDKSAAGWEYQAEVQKHGSQQDYSKGFGGKYGVQKDRQDKAAVGWE 575 +GFGGK+GVQ+DR D+SA G+EYQ + +KH SQ+DYS GFGGKYGVQ DR DK+AVG++ Sbjct: 127 VKGFGGKFGVQTDRVDQSAVGFEYQGKTEKHASQKDYSSGFGGKYGVQADRVDKSAVGFD 186 Query: 576 HKAELEKHSSQKDYSQGFGGKFGVQKERQDKSAVGWEHKEETEKHGSQQDYSKGFGGKYG 755 ++ + EKH SQKDYS+GFGGK+GV K++ DKSAVG+E++ +TEKH SQ+DY KGFGGK+G Sbjct: 187 YQGKTEKHESQKDYSKGFGGKYGVDKDKVDKSAVGFEYQGKTEKHESQKDYVKGFGGKFG 246 Query: 756 VQTDRQDKCAVGWEHKEELGKHASQQDYSQGFGGKFGVQKERQDKSAVGWEHKEETEKHG 935 VQTDRQDKCA+GW+H+E++ H SQ+DY GFGGKFGVQ ERQD SAVG+++KE+ KH Sbjct: 247 VQTDRQDKCALGWDHQEKVQLHESQKDYKSGFGGKFGVQTERQDPSAVGFDYKEKLAKHE 306 Query: 936 SQQDYSKGFGGKYGVQTDRQDKSAVGWE---------------------------HKEEV 1034 SQQDYSKGFGGKYGVQ DR DK+A +E + E + Sbjct: 307 SQQDYSKGFGGKYGVQKDRMDKNAATFEDIEKPTSTYQKTKPVERVANKTSSIRANLENL 366 Query: 1035 GKHSSQTDQSKGFGGKF-GVQSDRQDKSALGWDTEQVSKNVSHQNEPDVPTA------TV 1193 K Q D+ K + + ++Q++ E+ +K Q P PT T Sbjct: 367 AKEKEQEDRRKAEAERAQRMAREKQEQEEARRKLEEQAK-AKKQTPPPSPTTQPAEPKTP 425 Query: 1194 TKKIEQLSVKEDIRIEEQSSNDPV----EPVAGYG------EEHHSHDIEETEPQHNLET 1343 + + Q +V D ++S+ EP +GY +E S E EP+ E Sbjct: 426 SSPVYQDAVSYDAESAYKNSSTTYSAEHEPESGYKTTGSDYQEAVSQREAEYEPETVYEV 485 Query: 1344 NQSSDE---------------GLIAVALFDFDGEEEDEVNFAAGDTITHIEKIDSGWWVG 1478 + D G+ A+AL+D+ +DE++F D IT+IE ID GWW G Sbjct: 486 AGAGDHYQAEENTYDEYENELGITAIALYDYQAAGDDEISFDPDDIITNIEMIDDGWWRG 545 Query: 1479 VINGRKGMFPSNYVEINE 1532 V GR G+FP+NYVE+ + Sbjct: 546 VCKGRYGLFPANYVELRQ 563
>sp|Q60598|SRC8_MOUSE Src substrate cortactin Length = 546 Score = 451 bits (1159), Expect = e-126 Identities = 237/547 (43%), Positives = 327/547 (59%), Gaps = 50/547 (9%) Frame = +3 Query: 42 MWKATAGSXXXXXXXXXXXXXXXXXXXXFQNDVTEKEQRWGSKTVSGSGRTDFIDLSALK 221 MWKA+AG F NDV+EKEQRWG+KTV GSG + I++ L+ Sbjct: 1 MWKASAGHAVSITQDDGGADDWETDPD-FVNDVSEKEQRWGAKTVQGSGHQEHINIHKLR 59 Query: 222 ESVKKSDADNKKIAYDRSSKPSYGYGGSFGVQKDRMDKCAVGNDYVAAVDKHTSQKDYSQ 401 E+V + K+ + K S+GYGG FGV++DRMD+ AVG++Y + + KH SQ D + Sbjct: 60 ENVFQEHQTLKEKELETGPKASHGYGGKFGVEQDRMDRSAVGHEYQSKLSKHCSQVDSVR 119 Query: 402 GFGGKYGVQSDRKDKSAAGWEYQAEVQKHGSQQDYSKGFGGKYGVQKDRQDKAAVGWEHK 581 GFGGK+GVQ DR D+SA G+EYQ + +KH SQ+DYS GFGGKYGVQ DR DK+AVG++++ Sbjct: 120 GFGGKFGVQMDRVDQSAVGFEYQGKTEKHASQKDYSSGFGGKYGVQADRVDKSAVGFDYQ 179 Query: 582 AELEKHSSQKDYSQGFGGKFGVQKERQDKSAVGWEHKEETEKHGSQQDYSKGFGGKYGVQ 761 + EKH SQKDYS+GFGGK+G+ K++ DKSAVG+E++ +TEKH SQ+DY KGFGGK+GVQ Sbjct: 180 GKTEKHESQKDYSKGFGGKYGIDKDKVDKSAVGFEYQGKTEKHESQKDYVKGFGGKFGVQ 239 Query: 762 TDRQDKCAVGWEHKEELGKHASQQDYSQGFGGKFGVQKERQDKSAVGWEHKEETEKHGSQ 941 TDRQDKCA+GW+H+E+L H SQ+DY GFGGKFGVQ ERQD SAVG+++KE KH Q Sbjct: 240 TDRQDKCALGWDHQEKLQLHESQKDYKTGFGGKFGVQSERQDSSAVGFDYKERLAKHEPQ 299 Query: 942 QDYSKGFGGKYGVQTDRQDKSAVGWEHKEEVGKHSSQTDQSKGFGGKF--------GVQS 1097 QDY+KGFGGKYGVQ DR DK+A +E +V +T + K + Sbjct: 300 QDYAKGFGGKYGVQKDRMDKNASTFEEVVQVPSAYQKTVPIEAVTSKTSNIRANFENLAK 359 Query: 1098 DRQDKSALGWDTEQVSKNVSHQNEPD-----VPTATVTKKIEQLSVKEDIRIEEQSSNDP 1262 +R+ + + E+ + + E + + KK + IE++ + P Sbjct: 360 EREQEDRRKAEAERAQRMAKERQEQEEARRKLEEQARAKKQTPPASPSPQPIEDRPPSSP 419 Query: 1263 VEPVAGYGEEHHSHDIEETEPQHNLET--------------------------------- 1343 + A + S+ E EP++++E Sbjct: 420 IYEDAAPFKAEPSYRGSEPEPEYSIEAAGIPEAGSQQGLTYTSEPVYETTEAPGHYQAED 479 Query: 1344 ----NQSSDEGLIAVALFDFDGEEEDEVNFAAGDTITHIEKIDSGWWVGVINGRKGMFPS 1511 SD G+ A+AL+D+ +DE++F D IT+IE ID GWW GV GR G+FP+ Sbjct: 480 DTYDGYESDLGITAIALYDYQAAGDDEISFDPDDIITNIEMIDDGWWRGVCKGRYGLFPA 539 Query: 1512 NYVEINE 1532 NYVE+ + Sbjct: 540 NYVELRQ 546
>sp|Q14247|SRC8_HUMAN Src substrate cortactin (Amplaxin) (Oncogene EMS1) Length = 550 Score = 449 bits (1154), Expect = e-125 Identities = 247/552 (44%), Positives = 331/552 (59%), Gaps = 55/552 (9%) Frame = +3 Query: 42 MWKATAGSXXXXXXXXXXXXXXXXXXXXFQNDVTEKEQRWGSKTVSGSGRTDFIDLSALK 221 MWKA+AG F NDV+EKEQRWG+KTV GSG + I++ L+ Sbjct: 1 MWKASAGHAVSIAQDDAGADDWETDPD-FVNDVSEKEQRWGAKTVQGSGHQEHINIHKLR 59 Query: 222 ESVKKSDADNKKIAYDRSSKPSYGYGGSFGVQKDRMDKCAVGNDYVAAVDKHTSQKDYSQ 401 E+V + K+ + K S+GYGG FGV++DRMDK AVG++Y + + KH SQ D + Sbjct: 60 ENVFQEHQTLKEKELETGPKASHGYGGKFGVEQDRMDKSAVGHEYQSKLSKHCSQVDSVR 119 Query: 402 GFGGKYGVQSDRKDKSAAGWEYQAEVQKHGSQQDYSKGFGGKYGVQKDRQDKAAVGWEHK 581 GFGGK+GVQ DR D+SA G+EYQ + +KH SQ+DYS GFGGKYGVQ DR DK+AVG++++ Sbjct: 120 GFGGKFGVQMDRVDQSAVGFEYQGKTEKHASQKDYSSGFGGKYGVQADRVDKSAVGFDYQ 179 Query: 582 AELEKHSSQKDYSQGFGGKFGVQKERQDKSAVGWEHKEETEKHGSQQDYSKGFGGKYGVQ 761 + EKH SQ+DYS+GFGGK+G+ K++ DKSAVG+E++ +TEKH SQ+DY KGFGGK+GVQ Sbjct: 180 GKTEKHESQRDYSKGFGGKYGIDKDKVDKSAVGFEYQGKTEKHESQKDYVKGFGGKFGVQ 239 Query: 762 TDRQDKCAVGWEHKEELGKHASQQDYSQGFGGKFGVQKERQDKSAVGWEHKEETEKHGSQ 941 TDRQDKCA+GW+H+E+L H SQ+DY GFGGKFGVQ ERQD +AVG+++KE+ KH SQ Sbjct: 240 TDRQDKCALGWDHQEKLQLHESQKDYKTGFGGKFGVQSERQDSAAVGFDYKEKLAKHESQ 299 Query: 942 QDYSKGFGGKYGVQTDRQDKSAVGWE---------------------------HKEEVGK 1040 QDYSKGFGGKYGVQ DR DK+A +E + E + K Sbjct: 300 QDYSKGFGGKYGVQKDRMDKNASTFEDVTQVSSAYQKTVPVEAVTSKTSNIRANFENLAK 359 Query: 1041 HSSQTDQSKGFGGKF-GVQSDRQDKSALGWDTEQVSKNVSHQNEPDVPTATVTKK----- 1202 Q D+ K + + +RQ++ E+ ++ Q P P T++ Sbjct: 360 EKEQEDRRKAEAERAQRMAKERQEQEEARRKLEEQAR-AKTQTPPVSPAPQPTEERLPSS 418 Query: 1203 ---IEQLSVKEDIRIEEQSSNDPVEPV-----AGYGEEHHSHDI--------EETE-PQH 1331 + S K ++ S EPV A Y E + E E P H Sbjct: 419 PVYEDAASFKAELSYRGPVSGTEPEPVYSMEAADYREASSQQGLAYATEAVYESAEAPGH 478 Query: 1332 NLETNQSSDE-----GLIAVALFDFDGEEEDEVNFAAGDTITHIEKIDSGWWVGVINGRK 1496 + + DE G AVAL+D+ +DE++F D IT+IE ID GWW GV GR Sbjct: 479 YPAEDSTYDEYENDLGYTAVALYDYQAAGDDEISFDPDDIITNIEMIDDGWWRGVCKGRY 538 Query: 1497 GMFPSNYVEINE 1532 G+FP+NYVE+ + Sbjct: 539 GLFPANYVELRQ 550
>sp|P14317|HCLS1_HUMAN Hematopoietic lineage cell-specific protein (Hematopoietic cell-specific LYN substrate 1) (LckBP1) (p75) Length = 486 Score = 290 bits (741), Expect = 1e-77 Identities = 181/499 (36%), Positives = 266/499 (53%), Gaps = 30/499 (6%) Frame = +3 Query: 126 FQNDVTEKEQRWGSKTVSGSGRTDFIDLSALKESVKKSDADNKKIAYDRSSKPSYGYGGS 305 F ND++EKEQRWG+KT+ GSGRT+ I++ L+ V + +K + K S+GYGG Sbjct: 27 FVNDISEKEQRWGAKTIEGSGRTEHINIHQLRNKVSEEHDVLRKKEMESGPKASHGYGGR 86 Query: 306 FGVQKDRMDKCAVGNDYVAAVDKHTSQKDYSQGFGGKYGVQSDRKDKSAAGWEYQAEVQK 485 FGV++DRMDK AVG++YVA V+KH+SQ D ++GFGGKYGV+ DR DKSA G++Y+ EV+K Sbjct: 87 FGVERDRMDKSAVGHEYVAEVEKHSSQTDAAKGFGGKYGVERDRADKSAVGFDYKGEVEK 146 Query: 486 HGSQQDYSKGFGGKYGVQKDRQDKAAVGWEHKAELEKHSSQKDYSQGFGGKFGVQKERQD 665 H SQ+DYS+GFGG+YGV+KD+ DKAA+G+++K E EKH SQ+DY++GFGG++G+QK+R D Sbjct: 147 HTSQKDYSRGFGGRYGVEKDKWDKAALGYDYKGETEKHESQRDYAKGFGGQYGIQKDRVD 206 Query: 666 KSAVGWEHKE-ETEKHGSQQDYSKGFGGKYGVQTDRQDKCAVGWEHKEELGKHASQQDYS 842 KSAVG+ E T + G G++ + EE K ++ Sbjct: 207 KSAVGFNEMEAPTTAYKKTTPIEAASSGARGLKAKFESMA-------EEKRKREEEEKAQ 259 Query: 843 QGFGGKFGVQKERQDKSAVGWEHKEETEKHGSQQDYSKGFGGKYGVQTDRQDK-SAVGWE 1019 Q V + +Q++ AV E + + ++ + V K S+ W Sbjct: 260 Q-------VARRQQERKAVTKRSPEAPQPVIAMEEPA--------VPAPLPKKISSEAW- 303 Query: 1020 HKEEVGKHSSQTDQSKGFGGKFGVQSDRQDKSALGWDTEQVSKNVSHQNEPDVPTATVTK 1199 VG S + V++ R+ L + + + NE P A + Sbjct: 304 --PPVGTPPSSESEP--------VRTSREHPVPL----LPIRQTLPEDNEE--PPALPPR 347 Query: 1200 KIEQLSVKEDIRIEEQSSNDP---VEPVAGYG-----EEHHSHDIEETEPQHNLETNQSS 1355 +E L V+E+ E + +P EP Y + H D E + + LE SS Sbjct: 348 TLEGLQVEEEPVYEAEPEPEPEPEPEPENDYEDVEEMDRHEQEDEPEGDYEEVLEPEDSS 407 Query: 1356 DE--------------------GLIAVALFDFDGEEEDEVNFAAGDTITHIEKIDSGWWV 1475 G+ AVAL+D+ GE DE++F D IT IE +D GWW Sbjct: 408 FSSALAGSSGCPAGAGAGAVALGISAVALYDYQGEGSDELSFDPDDVITDIEMVDEGWWR 467 Query: 1476 GVINGRKGMFPSNYVEINE 1532 G +G G+FP+NYV++ E Sbjct: 468 GRCHGHFGLFPANYVKLLE 486
>sp|P49710|HCLS1_MOUSE Hematopoietic lineage cell-specific protein (Hematopoietic cell-specific LYN substrate 1) (LckBP1) Length = 486 Score = 287 bits (734), Expect = 7e-77 Identities = 178/494 (36%), Positives = 263/494 (53%), Gaps = 27/494 (5%) Frame = +3 Query: 126 FQNDVTEKEQRWGSKTVSGSGRTDFIDLSALKESVKKSDADNKKIAYDRSSKPSYGYGGS 305 F ND++EKEQRWG+KT+ GSGRT+ I++ L+ V + KK + K S+GYGG Sbjct: 27 FVNDISEKEQRWGAKTIEGSGRTEHINIHQLRNKVSEEHDILKKKELESGPKASHGYGGQ 86 Query: 306 FGVQKDRMDKCAVGNDYVAAVDKHTSQKDYSQGFGGKYGVQSDRKDKSAAGWEYQAEVQK 485 FGV++DRMDK AVG++YVA V+KH+SQ D ++GFGGKYGV+ DR DKSA G++Y+ EV+K Sbjct: 87 FGVERDRMDKSAVGHEYVADVEKHSSQTDAARGFGGKYGVERDRADKSAVGFDYKGEVEK 146 Query: 486 HGSQQDYSKGFGGKYGVQKDRQDKAAVGWEHKAELEKHSSQKDYSQGFGGKFGVQKERQD 665 H SQ+DYS GFGG+YGV+KD++DKAA+G+++K E EKH SQ+DY++GFGG++G+QK+R D Sbjct: 147 HASQKDYSHGFGGRYGVEKDKRDKAALGYDYKGETEKHESQRDYAKGFGGQYGIQKDRVD 206 Query: 666 KSAVGWEHKE-ETEKHGSQQDYSKGFGGKYGVQTDRQDKCAVGWEHKEELGKHASQQDYS 842 KSAVG+ E T + G G++ + EE K ++ Sbjct: 207 KSAVGFNEMEAPTTAYKKTTPIEAASSGARGLKAKFESLA-------EEKRKREEEEKAQ 259 Query: 843 QGFGGKFGVQKERQDKSAVGWEHKEETEKHGSQQDYSKGFGGKYGVQTDRQDKSAVGWEH 1022 Q + +++Q++ AV +E + ++ + Q ++ S V W Sbjct: 260 Q-------MARQQQERKAVVKMSREVQQPSMPVEEPAA------PAQLPKKISSEV-WPP 305 Query: 1023 KEEVGKHSSQTDQSKGFGGKFGVQSDRQDKSALGWDTEQVSKNVSHQNEPDVPTATVTKK 1202 E SQ +S R++ T Q S +H + + P A + Sbjct: 306 AESHLPPESQPVRS------------RREYPVPSLPTRQ-SPLQNHLEDNEEPPALPPRT 352 Query: 1203 IEQLSVKEDIRIEEQSSNDPVEPVAGYGEEHHS---------------------HDIEET 1319 E L V E+ E +P EP Y E + D+ E Sbjct: 353 PEGLQVVEEPVYEAAPELEP-EPEPDYEPEPETEPDYEDVGELDRQDEDAEGDYEDVLEP 411 Query: 1320 EPQHNLE-----TNQSSDEGLIAVALFDFDGEEEDEVNFAAGDTITHIEKIDSGWWVGVI 1484 E +L + + G+ A+AL+D+ GE DE++F D IT IE +D GWW G Sbjct: 412 EDTPSLSYQAGPSAGAGGAGISAIALYDYQGEGSDELSFDPDDIITDIEMVDEGWWRGQC 471 Query: 1485 NGRKGMFPSNYVEI 1526 G G+FP+NYV++ Sbjct: 472 RGHFGLFPANYVKL 485
Score = 193 bits (491), Expect = 1e-48
Identities = 109/288 (37%), Positives = 167/288 (57%), Gaps = 29/288 (10%)
Frame = +3
Query: 444 KSAAGWEYQAEVQKHGSQQDYSKGFGGKYGVQKDR------------------QDKAAVG 569
KS G + V+ G D F ++ R Q + V
Sbjct: 3 KSVVGHDVSVSVETQGDDWDTDPDFVNDISEKEQRWGAKTIEGSGRTEHINIHQLRNKVS 62
Query: 570 WEH----KAELEKHSSQKDYSQGFGGKFGVQKERQDKSAVGWEHKEETEKHGSQQDYSKG 737
EH K ELE S S G+GG+FGV+++R DKSAVG E+ + EKH SQ D ++G
Sbjct: 63 EEHDILKKKELE---SGPKASHGYGGQFGVERDRMDKSAVGHEYVADVEKHSSQTDAARG 119
Query: 738 FGGKYGVQTDRQDKCAVGWEHKEELGKHASQQDYSQGFGGKFGVQKERQDKSAVGWEHKE 917
FGGKYGV+ DR DK AVG+++K E+ KHASQ+DYS GFGG++GV+K+++DK+A+G+++K
Sbjct: 120 FGGKYGVERDRADKSAVGFDYKGEVEKHASQKDYSHGFGGRYGVEKDKRDKAALGYDYKG 179
Query: 918 ETEKHGSQQDYSKGFGGKYGVQTDRQDKSAVGWEHKE-------EVGKHSSQTDQSKGFG 1076
ETEKH SQ+DY+KGFGG+YG+Q DR DKSAVG+ E + + + ++G
Sbjct: 180 ETEKHESQRDYAKGFGGQYGIQKDRVDKSAVGFNEMEAPTTAYKKTTPIEAASSGARGLK 239
Query: 1077 GKFGVQSDRQDKSALGWDTEQVSKNVSHQNEPDVPTATVTKKIEQLSV 1220
KF ++ + K + E+ ++ ++ Q + ++++++Q S+
Sbjct: 240 AKFESLAEEKRKR----EEEEKAQQMARQQQERKAVVKMSREVQQPSM 283
>sp|Q6XPR3|RPTN_HUMAN Repetin Length = 784 Score = 67.0 bits (162), Expect = 2e-10 Identities = 89/337 (26%), Positives = 147/337 (43%), Gaps = 23/337 (6%) Frame = +3 Query: 234 KSDADNKKIAYDRSSKPSYGYGGSFGVQKDRMDKCAVGNDYVAAVDKHTSQ-KDYSQGFG 410 +S+ +K ++D+S + S V A ++ A+++ +D G Sbjct: 184 QSERQDKDFSFDQSERQSQDSSSGKKVSHKSTSGQAKWQGHIFALNRCEKPIQDSHYGQS 243 Query: 411 GKYGVQSDRKDKSAAGWEYQAEVQKHGSQQDYSKGFGGKYGV-QKDRQDKAAVGWEHKAE 587 ++ QS+ + A Q QK GS S+ G + G Q DRQ +++ H + Sbjct: 244 ERHTQQSETLGQ--ASHFNQTNQQKSGSYCGQSERLGQELGCGQTDRQGQSS----HYGQ 297 Query: 588 LEKHSSQKDYSQGFGGKFGVQKERQDKSAVGWEHKEETEKHGSQQDYSK----GFGGKYG 755 ++ +D S +G Q +RQ +S+ H +T++ G YS+ G YG Sbjct: 298 TDR----QDQSYHYG-----QTDRQGQSS----HYSQTDRQGQSSHYSQPDRQGQSSHYG 344 Query: 756 VQTDRQDKCA-VGWEHKEELGKHASQQDYSQGFGGKFGV-----------QKERQDKSAV 899 Q DR+ +C +++ G H SQ + QG +G Q +RQD+S+ Sbjct: 345 -QMDRKGQCYHYDQTNRQGQGSHYSQPN-RQGQSSHYGQPDTQDQSSHYGQTDRQDQSS- 401 Query: 900 GWEHKEETEKHGSQQDYS----KGFGGKYGVQTDRQDKSA-VGWEHKEEVGKHSSQTDQS 1064 H +TE+ G YS +G G YG QTDRQ +S+ G ++ H QTD+ Sbjct: 402 ---HYGQTERQGQSSHYSQMDRQGQGSHYG-QTDRQGQSSHYGQPDRQGQNSHYGQTDR- 456 Query: 1065 KGFGGKFGVQSDRQDKSALGWDTEQVSKNVSHQNEPD 1175 +G +G Q+DRQ +S SH ++PD Sbjct: 457 QGQSSHYG-QTDRQGQS-------------SHYSQPD 479
>sp|Q925Q9|SH3K1_RAT SH3-domain kinase binding protein 1 (SH3 containing, expressed in tumorigenic astrocytes) (Regulator of ubiquitous kinase) (Ruk) Length = 709 Score = 67.0 bits (162), Expect = 2e-10 Identities = 31/67 (46%), Positives = 45/67 (67%), Gaps = 1/67 (1%) Frame = +3 Query: 1371 AVALFDFDGEEEDEVNFAAGDTITHIEKIDSGWWVGVINGRKGMFPSNYV-EINE*KNIY 1547 A+ FD+ + +DE+ + G+ IT+I K D GWW G INGR+G+FP N+V EI K++ Sbjct: 4 AIVEFDYQAQHDDELTISVGEVITNIRKEDGGWWEGQINGRRGLFPDNFVREIK--KDVK 61 Query: 1548 KHLLINR 1568 K LL N+ Sbjct: 62 KDLLSNK 68
Score = 55.8 bits (133), Expect = 4e-07
Identities = 22/51 (43%), Positives = 35/51 (68%), Gaps = 2/51 (3%)
Frame = +3
Query: 1380 LFDFDGEEEDEVNFAAGDTITHIEK--IDSGWWVGVINGRKGMFPSNYVEI 1526
+F ++ + +DE+ GD +T I K ID GWW G +NGR+G+FP N+V++
Sbjct: 319 IFPYEAQNDDELTIKEGDIVTLINKDCIDVGWWEGELNGRRGVFPDNFVKL 369
Score = 55.5 bits (132), Expect = 5e-07
Identities = 29/107 (27%), Positives = 55/107 (51%), Gaps = 5/107 (4%)
Frame = +3
Query: 1218 VKEDIRIEEQSSNDPVEPVAGYGEEHHSHDIEE----TEPQHNLETNQSSDEGLIAVAL- 1382
+K+D++ + S+ P +P+ HD+ + L TN+ + +
Sbjct: 56 IKKDVKKDLLSNKAPEKPM---------HDVSSGNSLLSSETILRTNKRGERRRRRCQVA 106
Query: 1383 FDFDGEEEDEVNFAAGDTITHIEKIDSGWWVGVINGRKGMFPSNYVE 1523
F + + +DE+ GD I + +++ GWW GV+NG+ GMFPSN+++
Sbjct: 107 FSYLPQNDDELELKVGDIIEVVGEVEEGWWEGVLNGKTGMFPSNFIK 153
>sp|Q8R550|SH3K1_MOUSE SH3-domain kinase binding protein 1 (SH3 containing, expressed in tumorigenic astrocytes) (Regulator of ubiquitous kinase) (Ruk) Length = 709 Score = 66.2 bits (160), Expect = 3e-10 Identities = 31/67 (46%), Positives = 45/67 (67%), Gaps = 1/67 (1%) Frame = +3 Query: 1371 AVALFDFDGEEEDEVNFAAGDTITHIEKIDSGWWVGVINGRKGMFPSNYV-EINE*KNIY 1547 A+ FD+ + +DE+ + G+ IT+I K D GWW G INGR+G+FP N+V EI K++ Sbjct: 4 AIVEFDYQAQHDDELTISVGEVITNIRKEDGGWWEGQINGRRGLFPDNFVREIK--KDMK 61 Query: 1548 KHLLINR 1568 K LL N+ Sbjct: 62 KDLLSNK 68
Score = 55.8 bits (133), Expect = 4e-07
Identities = 22/51 (43%), Positives = 35/51 (68%), Gaps = 2/51 (3%)
Frame = +3
Query: 1380 LFDFDGEEEDEVNFAAGDTITHIEK--IDSGWWVGVINGRKGMFPSNYVEI 1526
+F ++ + +DE+ GD +T I K ID GWW G +NGR+G+FP N+V++
Sbjct: 319 IFPYEAQNDDELTIKEGDIVTLINKDCIDVGWWEGELNGRRGVFPDNFVKL 369
Score = 54.7 bits (130), Expect = 8e-07
Identities = 29/107 (27%), Positives = 55/107 (51%), Gaps = 5/107 (4%)
Frame = +3
Query: 1218 VKEDIRIEEQSSNDPVEPVAGYGEEHHSHDIEE----TEPQHNLETNQSSDEGLIAVAL- 1382
+K+D++ + S+ P +P+ HD+ + L TN+ + +
Sbjct: 56 IKKDMKKDLLSNKAPEKPM---------HDVSSGNALLSSETILRTNKRGERRRRRCQVA 106
Query: 1383 FDFDGEEEDEVNFAAGDTITHIEKIDSGWWVGVINGRKGMFPSNYVE 1523
F + + +DE+ GD I + +++ GWW GV+NG+ GMFPSN+++
Sbjct: 107 FSYLPQNDDELELKVGDIIEVVGEVEEGWWEGVLNGKTGMFPSNFIK 153
>sp|Q14847|LASP1_HUMAN LIM and SH3 domain protein 1 (LASP-1) (MLN 50) Length = 261 Score = 65.9 bits (159), Expect = 3e-10 Identities = 58/248 (23%), Positives = 96/248 (38%), Gaps = 3/248 (1%) Frame = +3 Query: 789 GWEHKEELGKHASQQDYSQGFGGKFGVQKERQDKSAVGWEHKEETEKHGSQQDYSKGFGG 968 G+E K H +Q ++ ++ ++Q + +KEE EK+ KG G Sbjct: 46 GYEKKPYCNAHYPKQSFTMVADTPENLRLKQQSELQSQVRYKEEFEKN-------KGKGF 98 Query: 969 KYGVQTDRQDKSAVGWEHKEEVGKHSSQTDQSKGFGGKFGVQSDRQDKSALGWDTEQVSK 1148 T + + + H G G G++ +R+D D + Sbjct: 99 SVVADTPELQRIKKTQDQISNIKYHEEFEKSRMGPSGGEGMEPERRDSQ----DGSSYRR 154 Query: 1149 NVSHQNEPDVPTATVTKKIEQLSVKEDIRIEEQSSNDPV-EPVAGYGEEHHSHDIEETEP 1325 + Q +PT+ + +Q PV + GY E I+ + P Sbjct: 155 PLEQQQPHHIPTSAP--------------VYQQPQQQPVAQSYGGYKEPAAPVSIQRSAP 200 Query: 1326 QHNLETNQSSDEGLIAVALFDFDGEEEDEVNFAAGDTITHIEKIDSGWWVGVI--NGRKG 1499 G A++D+ +EDEV+F GDTI ++++ID GW G + G G Sbjct: 201 ---------GGGGKRYRAVYDYSAADEDEVSFQDGDTIVNVQQIDDGWMYGTVERTGDTG 251 Query: 1500 MFPSNYVE 1523 M P+NYVE Sbjct: 252 MLPANYVE 259
>sp|Q99MZ8|LASP1_RAT LIM and SH3 domain protein 1 (LASP-1) Length = 263 Score = 65.5 bits (158), Expect = 4e-10 Identities = 62/251 (24%), Positives = 97/251 (38%), Gaps = 6/251 (2%) Frame = +3 Query: 789 GWEHKEELGKHASQQDYSQGFGGKFGVQKERQDKSAVGWEHKEETEKHGSQQDYSKGFGG 968 G+E K H +Q ++ ++ ++Q + +KEE EK+ KG G Sbjct: 46 GYEKKPYCNAHYPKQSFTMVADTPENLRLKQQSELQSQVRYKEEFEKN-------KGKGF 98 Query: 969 KYGVQTDRQDKSAVGWEHKEEVGKHSSQTDQSKGFGGKFGVQSDR---QDKSALGWDTEQ 1139 T + + + H G G G++ +R QD S+ TEQ Sbjct: 99 SVVADTPELQRIKKTQDQISNIKYHEEFEKSRMGPSGGEGIEPERREAQDSSSYRRPTEQ 158 Query: 1140 VSKNVSHQNEPDVPTATVTKKIEQLSVKEDIRIEEQSSNDPVEP-VAGYGEEHHSHDIEE 1316 H +PT+ + +Q V P GY E I+ Sbjct: 159 QQPQPHH-----IPTSAP--------------VYQQPQQQQVTPSYGGYKEPAAPVSIQR 199 Query: 1317 TEPQHNLETNQSSDEGLIAVALFDFDGEEEDEVNFAAGDTITHIEKIDSGWWVGVI--NG 1490 + P G A++D+ +EDEV+F GDTI ++++ID GW G + G Sbjct: 200 SAP---------GGGGKRYRAVYDYSAADEDEVSFQDGDTIVNVQQIDDGWMYGTVERTG 250 Query: 1491 RKGMFPSNYVE 1523 GM P+NYVE Sbjct: 251 DTGMLPANYVE 261
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 188,547,084
Number of Sequences: 369166
Number of extensions: 4189409
Number of successful extensions: 13937
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 11108
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 13647
length of database: 68,354,980
effective HSP length: 116
effective length of database: 46,925,720
effective search space used: 20647316800
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail