DrC_00398
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
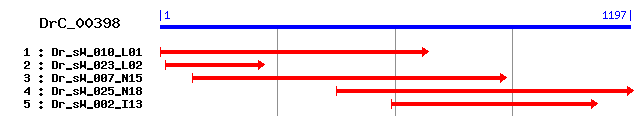
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00398 (1197 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|O02485|YDJ1_CAEEL Hypothetical protein ZK1073.1 in chrom... 151 4e-36 sp|Q62433|NDRG1_MOUSE NDRG1 protein (N-myc downstream regul... 122 3e-27 sp|Q92597|NDRG1_HUMAN NDRG1 protein (N-myc downstream regul... 119 1e-26 sp|Q9UN36|NDRG2_HUMAN NDRG2 protein (Syld709613 protein) 110 8e-24 sp|Q9UGV2|NDRG3_HUMAN NDRG3 protein 109 1e-23 sp|Q9QYG0|NDRG2_MOUSE NDRG2 protein (Ndr2 protein) 107 5e-23 sp|Q9QYF9|NDRG3_MOUSE NDRG3 protein (Ndr3 protein) 105 3e-22 sp|Q9ULP0|NDRG4_HUMAN NDRG4 protein (Brain development-rela... 105 3e-22 sp|Q8BTG7|NDRG4_MOUSE NDRG4 protein 104 4e-22 sp|Q9Z2L9|NDRG4_RAT NDRG4 protein (Brain development-relate... 102 2e-21
>sp|O02485|YDJ1_CAEEL Hypothetical protein ZK1073.1 in chromosome X Length = 325 Score = 151 bits (381), Expect = 4e-36 Identities = 100/334 (29%), Positives = 164/334 (49%), Gaps = 9/334 (2%) Frame = +2 Query: 2 LIMTTLNTKYCKTLTFYIEGTRSTN--EVAILTVHDIGCNHTQYVDFIKHSCMETLAPRC 175 L M + + C L Y++G + ILTVHDIG NH +V F+ H M T+ + Sbjct: 6 LQMVVVQAQNCGVLHVYVQGNLEERGGKTIILTVHDIGTNHKSFVRFVNHPSMATVKEKA 65 Query: 176 VWFNVDLPGQGINDPDLPSDFTYPTMQQIAEDLIECIGMLSISHLIVFGEGAGANILARM 355 ++ +V +PGQ N D DF PT+ I +DL + + I FGEG GANI+ R Sbjct: 66 IFLHVCVPGQEDNSADFFGDF--PTLDGIGDDLSAVLDKFEVKSAIAFGEGVGANIICRF 123 Query: 356 AMTGDERILGAVLLNCVGSTPNLVESLKDKIIGWKLSTIGMTSSTENYLVVHRFGVMNNL 535 AM RI+G VL++C +T ++E K+K++ +L M+ +YL+ H+FG + Sbjct: 124 AMGHPNRIMGIVLVHCTSTTAGIIEYCKEKVMNMRLENSIMSDGAWDYLLAHKFGGESKS 183 Query: 536 QSDEELSTMINNYKKFLRDTFNTKNLHGYVTSYMNRSNLIDKSMSIRCPM-----LFISG 700 + + Y + L+ T N KNL Y+ ++ R++L S +I + L ++G Sbjct: 184 RQE---------YLEELKATLNPKNLSKYLVAFTKRTDL---SSTIGTKLETVDALLVTG 231 Query: 701 SLANYADDVKCLYEDIQGFLKNDPDRKRHIEFMEIDNCANVIVERTDQVAESLQYFLQGQ 880 S A++ V ++ + K+ + +DN A+V+ E D++A SL +G Sbjct: 232 SKASHLHTVYTTHKSM---------NKKKTTLLVVDNVADVMQEAPDKLARSLILLCKGC 282 Query: 881 GLVG--SAPSRRMSSTVRPTGNRKISMEEYDSPK 976 G++ + P T+ SMEE D P+ Sbjct: 283 GVLSGVAIPGMERQRTL------SSSMEEADRPR 310
>sp|Q62433|NDRG1_MOUSE NDRG1 protein (N-myc downstream regulated gene 1 protein) (Protein Ndr1) Length = 394 Score = 122 bits (305), Expect = 3e-27 Identities = 93/335 (27%), Positives = 158/335 (47%), Gaps = 12/335 (3%) Frame = +2 Query: 59 GTRSTNEVAILTVHDIGCNH-TQYVDFIKHSCMETLAPRCVWFNVDLPGQGINDPDLPSD 235 GT N ILT HDIG NH T Y M+ + +VD PGQ P P Sbjct: 50 GTPKGNRPVILTYHDIGMNHKTCYNPLFNSEDMQEITQHFAVCHVDAPGQQDGAPSFPVG 109 Query: 236 FTYPTMQQIAEDLIECIGMLSISHLIVFGEGAGANILARMAMTGDERILGAVLLNCVGST 415 + YP+M Q+AE L + + +I G GAGA IL R A+ E + G VL+N Sbjct: 110 YMYPSMDQLAEMLPGVLHQFGLKSVIGMGTGAGAYILTRFALNNPEMVEGLVLMNVNPCA 169 Query: 416 PNLVESLKDKIIGWKLSTIGMTSSTENYLVVHRFG---VMNNLQSDEELSTMINNYKKFL 586 ++ KI GW T + + +V H FG + NN++ +++ Y++ + Sbjct: 170 EGWMDWAASKISGW-------TQALPDMVVSHLFGKEEIHNNVE-------VVHTYRQHI 215 Query: 587 RDTFNTKNLHGYVTSYMNRSNL-IDKSM------SIRCPMLFISGSLANYADDVKCLYED 745 + N NLH ++++Y +R +L I++ M +++CP L + G + D V Sbjct: 216 LNDMNPSNLHLFISAYNSRRDLEIERPMPGTHTVTLQCPALLVVGDNSPAVDAV------ 269 Query: 746 IQGFLKNDPDRKRHIEFMEIDNCANV-IVERTDQVAESLQYFLQGQGLVGSAPSRRMSST 922 ++ K DP + +++ +C + + + ++AE+ +YF+QG G + SA R+ + Sbjct: 270 VECNSKLDPTK---TTLLKMADCGGLPQISQPAKLAEAFKYFVQGMGYMPSASMTRLMRS 326 Query: 923 VRPTGNRKISMEEYDSPKGPSNITYNPRRKYSESE 1027 +G+ S+E + S+ + PR + SE Sbjct: 327 RTASGSSVTSLE---GTRSRSHTSEGPRSRSHTSE 358
>sp|Q92597|NDRG1_HUMAN NDRG1 protein (N-myc downstream regulated gene 1 protein) (Differentiation-related gene 1 protein) (DRG1) (Reducing agents and tunicamycin-responsive protein) (RTP) (Nickel-specific induction protein Cap43) (Rit42) Length = 394 Score = 119 bits (299), Expect = 1e-26 Identities = 92/332 (27%), Positives = 157/332 (47%), Gaps = 9/332 (2%) Frame = +2 Query: 59 GTRSTNEVAILTVHDIGCNH-TQYVDFIKHSCMETLAPRCVWFNVDLPGQGINDPDLPSD 235 GT N ILT HDIG NH T Y + M+ + +VD PGQ P+ Sbjct: 50 GTPKGNRPVILTYHDIGMNHKTCYNPLFNYEDMQEITQHFAVCHVDAPGQQDGAASFPAG 109 Query: 236 FTYPTMQQIAEDLIECIGMLSISHLIVFGEGAGANILARMAMTGDERILGAVLLNCVGST 415 + YP+M Q+AE L + + +I G GAGA IL R A+ E + G VL+N Sbjct: 110 YMYPSMDQLAEMLPGVLQQFGLKSIIGMGTGAGAYILTRFALNNPEMVEGLVLINVNPCA 169 Query: 416 PNLVESLKDKIIGWKLSTIGMTSSTENYLVVHRFGVMNNLQSDEELSTMINNYKKFLRDT 595 ++ KI GW T + + +V H FG +QS+ E +++ Y++ + + Sbjct: 170 EGWMDWAASKISGW-------TQALPDMVVSHLFG-KEEMQSNVE---VVHTYRQHIVND 218 Query: 596 FNTKNLHGYVTSYMNRSNL-IDKSM------SIRCPMLFISGSLANYADDVKCLYEDIQG 754 N NLH ++ +Y +R +L I++ M +++CP L + G + D V ++ Sbjct: 219 MNPGNLHLFINAYNSRRDLEIERPMPGTHTVTLQCPALLVVGDSSPAVDAV------VEC 272 Query: 755 FLKNDPDRKRHIEFMEIDNCANV-IVERTDQVAESLQYFLQGQGLVGSAPSRRMSSTVRP 931 K DP + +++ +C + + + ++AE+ +YF+QG G + SA R+ + Sbjct: 273 NSKLDPTK---TTLLKMADCGGLPQISQPAKLAEAFKYFVQGMGYMPSASMTRLMRSRTA 329 Query: 932 TGNRKISMEEYDSPKGPSNITYNPRRKYSESE 1027 +G+ S+ D + S+ + R + SE Sbjct: 330 SGS---SVTSLDGTRSRSHTSEGTRSRSHTSE 358
>sp|Q9UN36|NDRG2_HUMAN NDRG2 protein (Syld709613 protein) Length = 371 Score = 110 bits (275), Expect = 8e-24 Identities = 87/301 (28%), Positives = 144/301 (47%), Gaps = 8/301 (2%) Frame = +2 Query: 38 TLTFYIEGTRSTNEVAILTVHDIGCNHTQ-YVDFIKHSCMETLAPRCVWFNVDLPGQGIN 214 ++TF + GT AILT HD+G N+ + + M+ + V +VD PG Sbjct: 49 SVTFTVYGTPKPKRPAILTYHDVGLNYKSCFQPLFQFEDMQEIIQNFVRVHVDAPGMEEG 108 Query: 215 DPDLPSDFTYPTMQQIAEDLIECI-GMLSISHLIVFGEGAGANILARMAMTGDERILGAV 391 P P + YP++ Q+A D+I C+ L+ S +I G GAGA ILAR A+ + + G V Sbjct: 109 APVFPLGYQYPSLDQLA-DMIPCVLQYLNFSTIIGVGVGAGAYILARYALNHPDTVEGLV 167 Query: 392 LLNCVGSTPNLVESLKDKIIGWKLSTIGMTSSTENYLVVHRFGVMNNLQSDEEL---STM 562 L+N + ++ K+ G+TSS ++ H F S EEL S + Sbjct: 168 LINIDPNAKGWMDWAAHKL-------TGLTSSIPEMILGHLF-------SQEELSGNSEL 213 Query: 563 INNYKKFLRDTFNTKNLHGYVTSYMNRSNLIDK---SMSIRCPMLFISGSLANYADDVKC 733 I Y+ + N N+ Y SY NR +L + +++RCP++ + G A + D V Sbjct: 214 IQKYRNIITHAPNLDNIELYWNSYNNRRDLNFERGGDITLRCPVMLVVGDQAPHEDAV-- 271 Query: 734 LYEDIQGFLKNDPDRKRHIEFMEIDNCANVIVERTDQVAESLQYFLQGQGLVGSAPSRRM 913 ++ K DP + ++ D+ + + ++ E+ +YFLQG G + S+ R+ Sbjct: 272 ----VECNSKLDPTQTSFLKM--ADSGGQPQLTQPGKLTEAFKYFLQGMGYMASSCMTRL 325 Query: 914 S 916 S Sbjct: 326 S 326
>sp|Q9UGV2|NDRG3_HUMAN NDRG3 protein Length = 375 Score = 109 bits (273), Expect = 1e-23 Identities = 84/300 (28%), Positives = 138/300 (46%), Gaps = 12/300 (4%) Frame = +2 Query: 53 IEGTRSTNEVAILTVHDIGCNHTQYVD-FIKHSCMETLAPRCVWFNVDLPGQGINDPDLP 229 I G N ILT HDIG NH + F M+ + +VD PGQ P P Sbjct: 46 IRGLPKGNRPVILTYHDIGLNHKSCFNAFFNFEDMQEITQHFAVCHVDAPGQQEGAPSFP 105 Query: 230 SDFTYPTMQQIAEDLIECIGMLSISHLIVFGEGAGANILARMAMTGDERILGAVLLNCVG 409 + + YPTM ++AE L + LS+ +I G GAGA IL+R A+ E + G VL+N Sbjct: 106 TGYQYPTMDELAEMLPPVLTHLSLKSIIGIGVGAGAYILSRFALNHPELVEGLVLIN--- 162 Query: 410 STPNLVESLKDKIIGWKLSTI-GMTSSTENYLVVHRFGVMNNLQSDEELSTMINNYKKFL 586 V+ I W S + G+T++ + ++ H FG LQ++ +L I Y+ + Sbjct: 163 -----VDPCAKGWIDWAASKLSGLTTNVVDIILAHHFG-QEELQANLDL---IQTYRMHI 213 Query: 587 RDTFNTKNLHGYVTSYMNRSNL----------IDKSMSIRCPMLFISGSLANYADDVKCL 736 N NL ++ SY R +L +KS +++C L + G D+ + Sbjct: 214 AQDINQDNLQLFLNSYNGRRDLEIERPILGQNDNKSKTLKCSTLLVVG------DNSPAV 267 Query: 737 YEDIQGFLKNDPDRKRHIEFMEIDNCANVIVERTDQVAESLQYFLQGQGLVGSAPSRRMS 916 ++ + +P ++ + V+ + ++ E+ +YFLQG G + SA R++ Sbjct: 268 EAVVECNSRLNPINTTLLKMADCGGLPQVV--QPGKLTEAFKYFLQGMGYIPSASMTRLA 325
>sp|Q9QYG0|NDRG2_MOUSE NDRG2 protein (Ndr2 protein) Length = 371 Score = 107 bits (268), Expect = 5e-23 Identities = 85/301 (28%), Positives = 145/301 (48%), Gaps = 8/301 (2%) Frame = +2 Query: 38 TLTFYIEGTRSTNEVAILTVHDIGCNHTQ-YVDFIKHSCMETLAPRCVWFNVDLPGQGIN 214 ++TF + GT AI T HD+G N+ + + M+ + V +VD PG Sbjct: 49 SVTFTVYGTPKPKRPAIFTYHDVGLNYKSCFQPLFRFGDMQEIIQNFVRVHVDAPGMEEG 108 Query: 215 DPDLPSDFTYPTMQQIAEDLIECI-GMLSISHLIVFGEGAGANILARMAMTGDERILGAV 391 P P + YP++ Q+A D+I CI L+ S +I G GAGA IL+R A+ + + G V Sbjct: 109 APVFPLGYQYPSLDQLA-DMIPCILQYLNFSTIIGVGVGAGAYILSRYALNHPDTVEGLV 167 Query: 392 LLNCVGSTPNLVESLKDKIIGWKLSTIGMTSSTENYLVVHRFGVMNNLQSDEEL---STM 562 L+N + ++ K+ G+TSS + ++ H F S EEL S + Sbjct: 168 LINIDPNAKGWMDWAAHKL-------TGLTSSIPDMILGHLF-------SQEELSGNSEL 213 Query: 563 INNYKKFLRDTFNTKNLHGYVTSYMNRSNLIDK---SMSIRCPMLFISGSLANYADDVKC 733 I Y+ ++ N +N+ Y SY NR +L + +++CP++ + G A + D V Sbjct: 214 IQKYRGIIQHAPNLENIELYWNSYNNRRDLNFERGGETTLKCPVMLVVGDQAPHEDAV-- 271 Query: 734 LYEDIQGFLKNDPDRKRHIEFMEIDNCANVIVERTDQVAESLQYFLQGQGLVGSAPSRRM 913 ++ K DP + ++ D+ + + ++ E+ +YFLQG G + S+ R+ Sbjct: 272 ----VECNSKLDPTQTSFLKM--ADSGGQPQLTQPGKLTEAFKYFLQGMGYMASSCMTRL 325 Query: 914 S 916 S Sbjct: 326 S 326
>sp|Q9QYF9|NDRG3_MOUSE NDRG3 protein (Ndr3 protein) Length = 375 Score = 105 bits (262), Expect = 3e-22 Identities = 82/300 (27%), Positives = 137/300 (45%), Gaps = 12/300 (4%) Frame = +2 Query: 53 IEGTRSTNEVAILTVHDIGCNHTQ-YVDFIKHSCMETLAPRCVWFNVDLPGQGINDPDLP 229 I G N ILT HDIG NH + F M+ + +VD PGQ P P Sbjct: 46 IRGLPKGNRPVILTYHDIGLNHKSCFNTFFNFEDMQEITQHFAVCHVDAPGQQEAAPSFP 105 Query: 230 SDFTYPTMQQIAEDLIECIGMLSISHLIVFGEGAGANILARMAMTGDERILGAVLLNCVG 409 + + YPTM ++AE L + LS+ +I G GAGA IL+R A+ E + G VL+N Sbjct: 106 TGYQYPTMDELAEMLPPVLTHLSMKSIIGIGVGAGAYILSRFALNHPELVEGLVLIN--- 162 Query: 410 STPNLVESLKDKIIGWKLSTI-GMTSSTENYLVVHRFGVMNNLQSDEELSTMINNYKKFL 586 ++ I W S + G T++ + ++ H FG LQ++ +L I Y+ + Sbjct: 163 -----IDPCAKGWIDWAASKLSGFTTNIVDIILAHHFG-QEELQANLDL---IQTYRLHI 213 Query: 587 RDTFNTKNLHGYVTSYMNRSNL----------IDKSMSIRCPMLFISGSLANYADDVKCL 736 N +NL ++ SY R +L ++ +++C L + G D+ + Sbjct: 214 AQDINQENLQLFLGSYNGRRDLEIERPILGQNDNRLKTLKCSTLLVVG------DNSPAV 267 Query: 737 YEDIQGFLKNDPDRKRHIEFMEIDNCANVIVERTDQVAESLQYFLQGQGLVGSAPSRRMS 916 ++ + DP ++ + V+ + ++ E+ +YFLQG G + SA R++ Sbjct: 268 EAVVECNSRLDPINTTLLKMADCGGLPQVV--QPGKLTEAFKYFLQGMGYIPSASMTRLA 325
>sp|Q9ULP0|NDRG4_HUMAN NDRG4 protein (Brain development-related molecule 1) (Vascular smooth muscle cell associated protein 8) (SMAP-8) Length = 352 Score = 105 bits (261), Expect = 3e-22 Identities = 78/307 (25%), Positives = 139/307 (45%), Gaps = 11/307 (3%) Frame = +2 Query: 53 IEGTRSTNEVAILTVHDIGCNHTQ-YVDFIKHSCMETLAPRCVWFNVDLPGQGINDPDLP 229 I G+ N AILT HD+G NH + F M+ + V +VD PGQ + P Sbjct: 22 IRGSPKGNRPAILTYHDVGLNHKLCFNTFFNFEDMQEITKHFVVCHVDAPGQQVGASQFP 81 Query: 230 SDFTYPTMQQIAEDLIECIGMLSISHLIVFGEGAGANILARMAMTGDERILGAVLLNCVG 409 + +P+M+Q+A L + ++I G GAGA +LA+ A+ + + G VL+N Sbjct: 82 QGYQFPSMEQLAAMLPSVVQHFGFKYVIGIGVGAGAYVLAKFALIFPDLVEGLVLVNIDP 141 Query: 410 STPNLVESLKDKIIGWKLSTIGMTSSTENYLVVHRFGVMNNLQSDEEL---STMINNYKK 580 + ++ K+ G+TS+ + ++ H F S EEL + ++ +Y++ Sbjct: 142 NGKGWIDWAATKL-------SGLTSTLPDTVLSHLF-------SQEELVNNTELVQSYRQ 187 Query: 581 FLRDTFNTKNLHGYVTSY-------MNRSNLIDKSMSIRCPMLFISGSLANYADDVKCLY 739 + + N NL + Y +NR + + ++RCP++ + G A D V Sbjct: 188 QIGNVVNQANLQLFWNMYNSRRDLDINRPGTVPNAKTLRCPVMLVVGDNAPAEDGV---- 243 Query: 740 EDIQGFLKNDPDRKRHIEFMEIDNCANVIVERTDQVAESLQYFLQGQGLVGSAPSRRMSS 919 ++ K DP ++ + V + ++ E+ +YFLQG G + RR+S Sbjct: 244 --VECNSKLDPTTTTFLKMADSGGLPQ--VTQPGKLTEAFKYFLQGMGYIAYLKDRRLSG 299 Query: 920 TVRPTGN 940 P+ + Sbjct: 300 GAVPSAS 306
>sp|Q8BTG7|NDRG4_MOUSE NDRG4 protein Length = 352 Score = 104 bits (260), Expect = 4e-22 Identities = 77/307 (25%), Positives = 139/307 (45%), Gaps = 11/307 (3%) Frame = +2 Query: 53 IEGTRSTNEVAILTVHDIGCNHTQ-YVDFIKHSCMETLAPRCVWFNVDLPGQGINDPDLP 229 I G+ N AILT HD+G NH + F M+ + V +VD PGQ + P Sbjct: 22 IRGSPKGNRPAILTYHDVGLNHKLCFNTFFNFEDMQEITKHFVVCHVDAPGQQVGASQFP 81 Query: 230 SDFTYPTMQQIAEDLIECIGMLSISHLIVFGEGAGANILARMAMTGDERILGAVLLNCVG 409 + +P+M+Q+A L + ++I G GAGA +LA+ A+ + + G VL+N Sbjct: 82 QGYQFPSMEQLAAMLPSVVQHFGFKYVIGIGVGAGAYVLAKFALIFPDLVEGLVLMNIDP 141 Query: 410 STPNLVESLKDKIIGWKLSTIGMTSSTENYLVVHRFGVMNNLQSDEEL---STMINNYKK 580 + ++ K+ G+TS+ + ++ H F S EEL + ++ +Y++ Sbjct: 142 NGKGWIDWAATKL-------SGLTSTLPDTVLSHLF-------SQEELVNNTELVQSYRQ 187 Query: 581 FLRDTFNTKNLHGYVTSY-------MNRSNLIDKSMSIRCPMLFISGSLANYADDVKCLY 739 + + N NL + Y +NR + + ++RCP++ + G A + V Sbjct: 188 QISNVVNQANLQLFWNMYNSRRDLDINRPGTVPNAKTLRCPVMLVVGDNAPAEEGV---- 243 Query: 740 EDIQGFLKNDPDRKRHIEFMEIDNCANVIVERTDQVAESLQYFLQGQGLVGSAPSRRMSS 919 ++ K DP ++ + V + ++ E+ +YFLQG G + RR+S Sbjct: 244 --VECNSKLDPTTTTFLKMADSGGLPQ--VTQPGKLTEAFKYFLQGMGYIAHLKDRRLSG 299 Query: 920 TVRPTGN 940 P+ + Sbjct: 300 GAVPSAS 306
>sp|Q9Z2L9|NDRG4_RAT NDRG4 protein (Brain development-related molecule 1) Length = 352 Score = 102 bits (255), Expect = 2e-21 Identities = 77/307 (25%), Positives = 139/307 (45%), Gaps = 11/307 (3%) Frame = +2 Query: 53 IEGTRSTNEVAILTVHDIGCNHTQYVDFIKH-SCMETLAPRCVWFNVDLPGQGINDPDLP 229 I G+ N AILT HD+G NH + + + M+ + V +VD PGQ + P Sbjct: 22 IRGSPKGNRPAILTYHDVGLNHKLCFNTLFNLEDMQEITKHFVVCHVDAPGQQVGASQFP 81 Query: 230 SDFTYPTMQQIAEDLIECIGMLSISHLIVFGEGAGANILARMAMTGDERILGAVLLNCVG 409 + +P+M+Q+A L + ++I G GAGA +LA+ A+ + + G VL+N Sbjct: 82 QGYQFPSMEQLATMLPNVVQHFGFKYVIGIGVGAGAYVLAKFALIFPDLVEGLVLMNIDP 141 Query: 410 STPNLVESLKDKIIGWKLSTIGMTSSTENYLVVHRFGVMNNLQSDEEL---STMINNYKK 580 + ++ K+ G+TS+ + ++ H F S EEL + ++ +Y++ Sbjct: 142 NGKGWIDWAATKL-------SGLTSTLPDTVLSHLF-------SQEELVNNTELVQSYRQ 187 Query: 581 FLRDTFNTKNLHGYVTSY-------MNRSNLIDKSMSIRCPMLFISGSLANYADDVKCLY 739 + N NL + Y +NR + + ++RCP++ + G A D V Sbjct: 188 QISSVVNQANLQLFWNMYNSRRDLDINRPGTVPNAKTLRCPVMLVVGDNAPAEDGV---- 243 Query: 740 EDIQGFLKNDPDRKRHIEFMEIDNCANVIVERTDQVAESLQYFLQGQGLVGSAPSRRMSS 919 ++ K DP ++ + V + ++ E+ +YFLQG G + RR+S Sbjct: 244 --VECNSKLDPTTTTFLKMADSGGLPQ--VTQPGKLTEAFKYFLQGMGYIAHLKDRRLSG 299 Query: 920 TVRPTGN 940 P+ + Sbjct: 300 GAVPSAS 306
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 132,228,568
Number of Sequences: 369166
Number of extensions: 2720245
Number of successful extensions: 6455
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 6167
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6431
length of database: 68,354,980
effective HSP length: 113
effective length of database: 47,479,925
effective search space used: 13531778625
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail