DrC_00389
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00389 (1401 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P53590|SUCB2_PIG Succinyl-CoA ligase [GDP-forming] beta-... 479 e-135 sp|Q96I99|SUCB2_HUMAN Succinyl-CoA ligase [GDP-forming] bet... 477 e-134 sp|Q9Z2I8|SUCB2_MOUSE Succinyl-CoA ligase [GDP-forming] bet... 472 e-132 sp|P53589|SUCB2_CAEEL Probable succinyl-CoA ligase [GDP-for... 422 e-117 sp|Q9Z2I9|SUCB1_MOUSE Succinyl-CoA ligase [ADP-forming] bet... 383 e-106 sp|O97580|SUCB1_PIG Succinyl-CoA ligase [ADP-forming] beta-... 381 e-105 sp|Q9P567|SUCB_NEUCR Probable succinyl-CoA ligase [GDP-form... 380 e-105 sp|O82662|SUCB_ARATH Succinyl-CoA ligase [GDP-forming] beta... 380 e-105 sp|Q9P2R7|SUCB1_HUMAN Succinyl-CoA ligase [ADP-forming] bet... 377 e-104 sp|P53587|SUCB_NEOFR Succinyl-CoA ligase [GDP-forming] beta... 354 4e-97
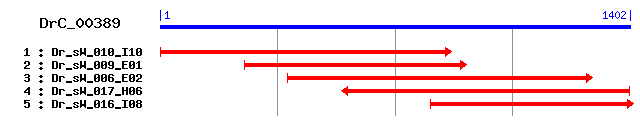
>sp|P53590|SUCB2_PIG Succinyl-CoA ligase [GDP-forming] beta-chain, mitochondrial precursor (Succinyl-CoA synthetase, betaG chain) (SCS-betaG) (GTP-specific succinyl-CoA synthetase beta subunit) Length = 433 Score = 479 bits (1233), Expect = e-135 Identities = 247/397 (62%), Positives = 302/397 (76%) Frame = +1 Query: 58 RNLNLQEYQSKGLMSKFGVNIQKFKVASTEQDAIDISKTFKVKEYVIKAQILAGGRGKGT 237 R LNLQEYQSK LMS GV +Q+F VA T +A++ +K KE V+KAQILAGGRGKG Sbjct: 37 RWLNLQEYQSKKLMSDNGVKVQRFFVADTANEALEAAKRLNAKEIVLKAQILAGGRGKGV 96 Query: 238 FSNGFKGGVHLTKNPSDVPNLVKSMLGQRLVTKQTSKDGVMVHKVMVAEALDIDRETYLA 417 FS+G KGGVHLTK+P V L K M+G L TKQT K+GV V+KVMVAEALDI RETYLA Sbjct: 97 FSSGLKGGVHLTKDPEVVGQLAKQMIGYNLATKQTPKEGVKVNKVMVAEALDISRETYLA 156 Query: 418 ILMDRTSGGLVVVSSPQGGMDIEEVAIKNPAAILKTKVDINKGITKSQCDEIAQHLLFTS 597 ILMDR+ G V+V SPQGG+DIEEVA NP I K ++DI +GI SQ +A++L F Sbjct: 157 ILMDRSCNGPVLVGSPQGGVDIEEVAASNPELIFKEQIDIIEGIKDSQAQRMAENLGFLG 216 Query: 598 KQQIDEVGKQLRGLFDLFLQHDCTQIEINPFGVTPGGQIVCFDAKLNFDDNAAFRQREIF 777 Q ++ Q++ L++LFL+ D TQ+E+NPFG TP GQ+VCFDAK+NFDDNA FRQ++IF Sbjct: 217 PLQ-NQAADQIKKLYNLFLKIDATQVEVNPFGETPEGQVVCFDAKINFDDNAEFRQKDIF 275 Query: 778 SLHDPVAEEQDpreIEAAQNGLNYIGLHGNIGCVVNGAGLAMATMDIVKLNGGTPANFLD 957 ++ D E +P E EAA+ L YIGL GNI C VNGAGLAMAT DI+ LNGG PANFLD Sbjct: 276 AMDD--KSENEPIENEAAKYDLKYIGLDGNIACFVNGAGLAMATCDIIFLNGGKPANFLD 333 Query: 958 LGGGVTASQVKQAFHILTGDPQVKSIFINIFGGIVNCGTIAEGLTAACRDVELKVPLIVR 1137 LGGGV SQV QAF +LT DP+V++I +NIFGGIVNC IA G+T ACR++ELKVPL+VR Sbjct: 334 LGGGVKESQVYQAFKLLTADPKVEAILVNIFGGIVNCAIIANGITKACRELELKVPLVVR 393 Query: 1138 LEGTNVDSARRILSNSGIPMKMSTDLDEAAKMAVHSV 1248 LEGTNV A+ IL+NSG+P+ + DL++AAK AV SV Sbjct: 394 LEGTNVHEAQNILTNSGLPITSAVDLEDAAKKAVASV 430
>sp|Q96I99|SUCB2_HUMAN Succinyl-CoA ligase [GDP-forming] beta-chain, mitochondrial precursor (Succinyl-CoA synthetase, betaG chain) (SCS-betaG) (GTP-specific succinyl-CoA synthetase beta subunit) Length = 432 Score = 477 bits (1227), Expect = e-134 Identities = 245/398 (61%), Positives = 303/398 (76%) Frame = +1 Query: 58 RNLNLQEYQSKGLMSKFGVNIQKFKVASTEQDAIDISKTFKVKEYVIKAQILAGGRGKGT 237 R LNLQEYQSK LMS GV +Q+F VA T +A++ +K KE V+KAQILAGGRGKG Sbjct: 36 RWLNLQEYQSKKLMSDNGVRVQRFFVADTANEALEAAKRLNAKEIVLKAQILAGGRGKGV 95 Query: 238 FSNGFKGGVHLTKNPSDVPNLVKSMLGQRLVTKQTSKDGVMVHKVMVAEALDIDRETYLA 417 F++G KGGVHLTK+P+ V L K M+G L TKQT K+GV V+KVMVAEALDI RETYLA Sbjct: 96 FNSGLKGGVHLTKDPNVVGQLAKQMIGYNLATKQTPKEGVKVNKVMVAEALDISRETYLA 155 Query: 418 ILMDRTSGGLVVVSSPQGGMDIEEVAIKNPAAILKTKVDINKGITKSQCDEIAQHLLFTS 597 ILMDR+ G V+V SPQGG+DIEEVA NP I K ++DI +GI SQ +A++L F Sbjct: 156 ILMDRSCNGPVLVGSPQGGVDIEEVAASNPELIFKEQIDIFEGIKDSQAQRMAENLGFVG 215 Query: 598 KQQIDEVGKQLRGLFDLFLQHDCTQIEINPFGVTPGGQIVCFDAKLNFDDNAAFRQREIF 777 + + Q+ L++LFL+ D TQ+E+NPFG TP GQ+VCFDAK+NFDDNA FRQ++IF Sbjct: 216 PLK-SQAADQITKLYNLFLKIDATQVEVNPFGETPEGQVVCFDAKINFDDNAEFRQKDIF 274 Query: 778 SLHDPVAEEQDpreIEAAQNGLNYIGLHGNIGCVVNGAGLAMATMDIVKLNGGTPANFLD 957 ++ D E +P E EAA+ L YIGL GNI C VNGAGLAMAT DI+ LNGG PANFLD Sbjct: 275 AMDD--KSENEPIENEAAKYDLKYIGLDGNIACFVNGAGLAMATCDIIFLNGGKPANFLD 332 Query: 958 LGGGVTASQVKQAFHILTGDPQVKSIFINIFGGIVNCGTIAEGLTAACRDVELKVPLIVR 1137 LGGGV +QV QAF +LT DP+V++I +NIFGGIVNC IA G+T ACR++ELKVPL+VR Sbjct: 333 LGGGVKEAQVYQAFKLLTADPKVEAILVNIFGGIVNCAIIANGITKACRELELKVPLVVR 392 Query: 1138 LEGTNVDSARRILSNSGIPMKMSTDLDEAAKMAVHSVA 1251 LEGTNV A++IL+NSG+P+ + DL++AAK AV SVA Sbjct: 393 LEGTNVQEAQKILNNSGLPITSAIDLEDAAKKAVASVA 430
>sp|Q9Z2I8|SUCB2_MOUSE Succinyl-CoA ligase [GDP-forming] beta-chain, mitochondrial precursor (Succinyl-CoA synthetase, betaG chain) (SCS-betaG) (GTP-specific succinyl-CoA synthetase beta subunit) Length = 404 Score = 472 bits (1214), Expect = e-132 Identities = 241/398 (60%), Positives = 303/398 (76%) Frame = +1 Query: 58 RNLNLQEYQSKGLMSKFGVNIQKFKVASTEQDAIDISKTFKVKEYVIKAQILAGGRGKGT 237 R LNLQEYQSK LMS+ GV +Q+F VA+T ++A++ +K KE V+KAQILAGGRGKG Sbjct: 8 RWLNLQEYQSKKLMSEHGVRVQRFFVANTAKEALEAAKRLNAKEIVLKAQILAGGRGKGV 67 Query: 238 FSNGFKGGVHLTKNPSDVPNLVKSMLGQRLVTKQTSKDGVMVHKVMVAEALDIDRETYLA 417 F++G KGGVHLTK+P V L + M+G L TKQT K+GV V+KVMVAEALDI RETYLA Sbjct: 68 FNSGLKGGVHLTKDPKVVGELAQQMIGYNLATKQTPKEGVKVNKVMVAEALDISRETYLA 127 Query: 418 ILMDRTSGGLVVVSSPQGGMDIEEVAIKNPAAILKTKVDINKGITKSQCDEIAQHLLFTS 597 ILMDR+ G V+V SPQGG+DIEEVA +P I K ++DI +GI SQ +A++L F Sbjct: 128 ILMDRSHNGPVIVGSPQGGVDIEEVAASSPELIFKEQIDIFEGIKDSQAQRMAENLGFLG 187 Query: 598 KQQIDEVGKQLRGLFDLFLQHDCTQIEINPFGVTPGGQIVCFDAKLNFDDNAAFRQREIF 777 + ++ Q+ L+ LFL+ D TQ+E+NPFG TP GQ+VCFDAK+NFDDNA FRQ++IF Sbjct: 188 SLK-NQAADQITKLYHLFLKIDATQVEVNPFGETPEGQVVCFDAKINFDDNAEFRQKDIF 246 Query: 778 SLHDPVAEEQDpreIEAAQNGLNYIGLHGNIGCVVNGAGLAMATMDIVKLNGGTPANFLD 957 ++ D E +P E EAA+ L YIGL GNI C VNGAGLAMAT DI+ LNGG PANFLD Sbjct: 247 AMDD--KSENEPIENEAARYDLKYIGLDGNIACFVNGAGLAMATCDIIFLNGGKPANFLD 304 Query: 958 LGGGVTASQVKQAFHILTGDPQVKSIFINIFGGIVNCGTIAEGLTAACRDVELKVPLIVR 1137 LGGGV +QV +AF +LT DP+V++I +NIFGGIVNC IA G+T ACR++ELKVPL+VR Sbjct: 305 LGGGVKEAQVYEAFKLLTSDPKVEAILVNIFGGIVNCAIIANGITKACRELELKVPLVVR 364 Query: 1138 LEGTNVDSARRILSNSGIPMKMSTDLDEAAKMAVHSVA 1251 LEGTNV A+ IL +SG+P+ + DL++AAK AV SVA Sbjct: 365 LEGTNVQEAQNILKSSGLPITSAVDLEDAAKKAVASVA 402
>sp|P53589|SUCB2_CAEEL Probable succinyl-CoA ligase [GDP-forming] beta-chain, mitochondrial precursor (Succinyl-CoA synthetase, beta chain) (SCS-beta) Length = 415 Score = 422 bits (1085), Expect = e-117 Identities = 221/401 (55%), Positives = 285/401 (71%), Gaps = 2/401 (0%) Frame = +1 Query: 52 QTRNLNLQEYQSKGLMSKFGVNIQKFKVASTEQDAIDISKTFKVKEYVIKAQILAGGRGK 231 Q R LNLQE+QSK ++ K G ++Q F VAS ++A + +F EYV+KAQILAGGRGK Sbjct: 16 QRRFLNLQEFQSKEILEKHGCSVQNFVVASNRKEAEEKWMSFGDHEYVVKAQILAGGRGK 75 Query: 232 GTFSNGFKG--GVHLTKNPSDVPNLVKSMLGQRLVTKQTSKDGVMVHKVMVAEALDIDRE 405 G F NG KG GV +TK + M+G+RLVTKQT+ +GV V KVM+AE +DI RE Sbjct: 76 GKFINGTKGIGGVFITKEKDAALEAIDEMIGKRLVTKQTTSEGVRVDKVMIAEGVDIKRE 135 Query: 406 TYLAILMDRTSGGLVVVSSPQGGMDIEEVAIKNPAAILKTKVDINKGITKSQCDEIAQHL 585 TYLA+LMDR S G VVV+SP GGMDIE VA K P I KT +DI G+T+ Q +IA+ L Sbjct: 136 TYLAVLMDRESNGPVVVASPDGGMDIEAVAEKTPERIFKTPIDIQMGMTEGQSLKIAKDL 195 Query: 586 LFTSKQQIDEVGKQLRGLFDLFLQHDCTQIEINPFGVTPGGQIVCFDAKLNFDDNAAFRQ 765 F K I ++++ L+DLF+ D TQ+EINP T G++ C DAK+NFDD+AA+RQ Sbjct: 196 QFEGKL-IGVAAQEIKRLYDLFIAVDATQVEINPLVETADGRVFCVDAKMNFDDSAAYRQ 254 Query: 766 REIFSLHDPVAEEQDpreIEAAQNGLNYIGLHGNIGCVVNGAGLAMATMDIVKLNGGTPA 945 +EIF+ EE Dpre++A Q LNYIG+ GNI C+VNGAGLAMATMD++KL+GG PA Sbjct: 255 KEIFAYE--TFEEHDpreVDAHQFNLNYIGMDGNIACLVNGAGLAMATMDLIKLHGGEPA 312 Query: 946 NFLDLGGGVTASQVKQAFHILTGDPQVKSIFINIFGGIVNCGTIAEGLTAACRDVELKVP 1125 NFLD+GG VT V A I+T DP+VK + INIFGGIVNC TIA G+ +A + L VP Sbjct: 313 NFLDVGGAVTEDAVFNAVRIITSDPRVKCVLINIFGGIVNCATIANGVVSAVNKIGLNVP 372 Query: 1126 LIVRLEGTNVDSARRILSNSGIPMKMSTDLDEAAKMAVHSV 1248 ++VRLEGTNVD+A++I+ SG+ + + +LDEAA AV S+ Sbjct: 373 MVVRLEGTNVDAAKQIMKKSGLKILTANNLDEAAAKAVSSL 413
>sp|Q9Z2I9|SUCB1_MOUSE Succinyl-CoA ligase [ADP-forming] beta-chain, mitochondrial precursor (Succinyl-CoA synthetase, betaA chain) (SCS-betaA) (ATP-specific succinyl-CoA synthetase beta subunit) Length = 463 Score = 383 bits (983), Expect = e-106 Identities = 192/397 (48%), Positives = 280/397 (70%) Frame = +1 Query: 49 KQTRNLNLQEYQSKGLMSKFGVNIQKFKVASTEQDAIDISKTFKVKEYVIKAQILAGGRG 228 +Q R L+L EY S L+ + GV++ K VA + +A I+K K+ VIKAQ+LAGGRG Sbjct: 48 QQQRTLSLHEYLSMELLQEAGVSVPKGFVAKSSDEAYAIAKKLGSKDVVIKAQVLAGGRG 107 Query: 229 KGTFSNGFKGGVHLTKNPSDVPNLVKSMLGQRLVTKQTSKDGVMVHKVMVAEALDIDRET 408 KGTF++G KGGV + +P + + M+GQ+L+TKQT + G + ++V+V E RE Sbjct: 108 KGTFTSGLKGGVKIVFSPEEAKAVSSQMIGQKLITKQTGEKGRICNQVLVCERKYPRREY 167 Query: 409 YLAILMDRTSGGLVVVSSPQGGMDIEEVAIKNPAAILKTKVDINKGITKSQCDEIAQHLL 588 Y AI M+R+ G V++ S QGG++IE+VA +NP AI+K +DI +GI K Q +AQ + Sbjct: 168 YFAITMERSFQGPVLIGSAQGGVNIEDVAAENPEAIVKEPIDIVEGIKKEQAVTLAQKMG 227 Query: 589 FTSKQQIDEVGKQLRGLFDLFLQHDCTQIEINPFGVTPGGQIVCFDAKLNFDDNAAFRQR 768 F S +D + + L++LFL++D T +EINP G+++C DAK+NFD N+A+RQ+ Sbjct: 228 FPSNI-VDSAAENMIKLYNLFLKYDATMVEINPMVEDSDGKVLCMDAKINFDSNSAYRQK 286 Query: 769 EIFSLHDPVAEEQDpreIEAAQNGLNYIGLHGNIGCVVNGAGLAMATMDIVKLNGGTPAN 948 +IF L D ++D R+ EAA +NYIGL G+IGC+VNGAGLAMATMDI+KL+GGTPAN Sbjct: 287 KIFDLQD--WSQEDERDKEAANADINYIGLDGSIGCLVNGAGLAMATMDIIKLHGGTPAN 344 Query: 949 FLDLGGGVTASQVKQAFHILTGDPQVKSIFINIFGGIVNCGTIAEGLTAACRDVELKVPL 1128 FLD+GGG T QV +AF ++T D +V++I +NIFGGI+ C IA+G+ A +D+E+++P+ Sbjct: 345 FLDVGGGATVQQVTEAFKLITSDKKVQAILVNIFGGIMRCDVIAQGIVMAVKDLEIRIPV 404 Query: 1129 IVRLEGTNVDSARRILSNSGIPMKMSTDLDEAAKMAV 1239 +VRL+GT VD A+ ++++SG+ + DLDEAAKM V Sbjct: 405 VVRLQGTRVDDAKALIADSGLKILACDDLDEAAKMVV 441
>sp|O97580|SUCB1_PIG Succinyl-CoA ligase [ADP-forming] beta-chain, mitochondrial precursor (Succinyl-CoA synthetase, betaA chain) (SCS-betaA) (ATP-specific succinyl-CoA synthetase beta subunit) Length = 425 Score = 381 bits (979), Expect = e-105 Identities = 195/397 (49%), Positives = 275/397 (69%) Frame = +1 Query: 49 KQTRNLNLQEYQSKGLMSKFGVNIQKFKVASTEQDAIDISKTFKVKEYVIKAQILAGGRG 228 +Q RNL+L EY S L+ + GV+I K VA + +A I+K K+ VIKAQ+LAGGRG Sbjct: 10 QQQRNLSLHEYMSMELLQEAGVSIPKGHVAKSPDEAYAIAKKLGSKDVVIKAQVLAGGRG 69 Query: 229 KGTFSNGFKGGVHLTKNPSDVPNLVKSMLGQRLVTKQTSKDGVMVHKVMVAEALDIDRET 408 KGTF +G KGGV + +P + + M+G++L TKQT + G + ++V+V E RE Sbjct: 70 KGTFESGLKGGVKIVFSPEEAKAVSSQMIGKKLFTKQTGEKGRICNQVLVCERRYPRREY 129 Query: 409 YLAILMDRTSGGLVVVSSPQGGMDIEEVAIKNPAAILKTKVDINKGITKSQCDEIAQHLL 588 Y AI M+R+ G V++ S QGG++IE+VA + P AI+K +DI +GI K Q +AQ + Sbjct: 130 YFAITMERSFQGPVLIGSSQGGVNIEDVAAETPEAIVKEPIDIVEGIKKEQAVRLAQKMG 189 Query: 589 FTSKQQIDEVGKQLRGLFDLFLQHDCTQIEINPFGVTPGGQIVCFDAKLNFDDNAAFRQR 768 F +D + + L++LFL++D T +EINP G ++C DAK+NFD N+A+RQ+ Sbjct: 190 FPPSI-VDSAAENMIKLYNLFLKYDATMVEINPMVEDSDGAVLCMDAKINFDSNSAYRQK 248 Query: 769 EIFSLHDPVAEEQDpreIEAAQNGLNYIGLHGNIGCVVNGAGLAMATMDIVKLNGGTPAN 948 +IF L D E D R+ +AA+ LNYIGL GNIGC+VNGAGLAMATMDI+KL+GGTPAN Sbjct: 249 KIFDLQDWTQE--DERDKDAAKANLNYIGLDGNIGCLVNGAGLAMATMDIIKLHGGTPAN 306 Query: 949 FLDLGGGVTASQVKQAFHILTGDPQVKSIFINIFGGIVNCGTIAEGLTAACRDVELKVPL 1128 FLD+GGG T QV +AF ++T D +V SI +NIFGGI+ C IA+G+ A +D+E+K+P+ Sbjct: 307 FLDVGGGATVHQVTEAFKLITSDKKVLSILVNIFGGIMRCDVIAQGIVMAVKDLEIKIPV 366 Query: 1129 IVRLEGTNVDSARRILSNSGIPMKMSTDLDEAAKMAV 1239 +VRL+GT VD A+ ++++SG+ + DLDEAAKM V Sbjct: 367 VVRLQGTRVDDAKALIADSGLKILACDDLDEAAKMVV 403
>sp|Q9P567|SUCB_NEUCR Probable succinyl-CoA ligase [GDP-forming] beta-chain, mitochondrial precursor (Succinyl-CoA synthetase, beta chain) (SCS-beta) Length = 447 Score = 380 bits (976), Expect = e-105 Identities = 196/397 (49%), Positives = 270/397 (68%) Frame = +1 Query: 49 KQTRNLNLQEYQSKGLMSKFGVNIQKFKVASTEQDAIDISKTFKVKEYVIKAQILAGGRG 228 +Q R L++ EY S L+ ++G+ + K VA T +A I+K ++ VIKAQ+LAGGRG Sbjct: 32 QQRRALSIHEYLSADLLRQYGIGVPKGDVARTGAEAEAIAKQIGGEDMVIKAQVLAGGRG 91 Query: 229 KGTFSNGFKGGVHLTKNPSDVPNLVKSMLGQRLVTKQTSKDGVMVHKVMVAEALDIDRET 408 KGTF NG KGGV + +P++ + M+G +L+TKQT G + V + E RE Sbjct: 92 KGTFDNGLKGGVRVIYSPTEAKMFAEQMIGHKLITKQTGAQGRLCSAVYICERKFARREF 151 Query: 409 YLAILMDRTSGGLVVVSSPQGGMDIEEVAIKNPAAILKTKVDINKGITKSQCDEIAQHLL 588 YLA+LMDR S G V+VSS QGGMDIE VA +NP AI T +DIN G+T +IA L Sbjct: 152 YLAVLMDRASQGPVIVSSSQGGMDIEGVAKENPEAIHTTYIDINVGVTDEMARDIATKLG 211 Query: 589 FTSKQQIDEVGKQLRGLFDLFLQHDCTQIEINPFGVTPGGQIVCFDAKLNFDDNAAFRQR 768 F S+Q ++E ++ L+ +F + D TQIEINP T +++ DAK FDDNA FRQ Sbjct: 212 F-SEQCVEEAKDTIQKLYKIFCEKDATQIEINPLSETSDHKVMAMDAKFGFDDNADFRQP 270 Query: 769 EIFSLHDPVAEEQDpreIEAAQNGLNYIGLHGNIGCVVNGAGLAMATMDIVKLNGGTPAN 948 +IF L D E DP E+ AAQ GLN+I L G+IGC+VNGAGLAMATMDI+KLNGG PAN Sbjct: 271 DIFKLRDTTQE--DPDEVRAAQAGLNFIKLDGDIGCLVNGAGLAMATMDIIKLNGGQPAN 328 Query: 949 FLDLGGGVTASQVKQAFHILTGDPQVKSIFINIFGGIVNCGTIAEGLTAACRDVELKVPL 1128 FLD+GGG T + +++AF ++T DP+V +IF+NIFGGIV C IA GL + ++LK+P+ Sbjct: 329 FLDVGGGATPAAIREAFELITSDPKVTAIFVNIFGGIVRCDAIAHGLINTVKSLDLKIPI 388 Query: 1129 IVRLEGTNVDSARRILSNSGIPMKMSTDLDEAAKMAV 1239 I RL+GTN+++AR+++++SG+ + DL AA+ +V Sbjct: 389 IARLQGTNMEAARQLINDSGMKIFSIDDLQSAAEKSV 425
>sp|O82662|SUCB_ARATH Succinyl-CoA ligase [GDP-forming] beta-chain, mitochondrial precursor (Succinyl-CoA synthetase, beta chain) (SCS-beta) Length = 421 Score = 380 bits (975), Expect = e-105 Identities = 201/403 (49%), Positives = 272/403 (67%), Gaps = 2/403 (0%) Frame = +1 Query: 49 KQTRNLNLQEYQSKGLMSKFGVNIQKFKVAST-EQDAIDISKTF-KVKEYVIKAQILAGG 222 +Q R LN+ EYQ LM K+GVN+ K AS+ E+ I F E V+K+QILAGG Sbjct: 22 QQLRRLNIHEYQGAELMGKYGVNVPKGVAASSLEEVKKAIQDVFPNESELVVKSQILAGG 81 Query: 223 RGKGTFSNGFKGGVHLTKNPSDVPNLVKSMLGQRLVTKQTSKDGVMVHKVMVAEALDIDR 402 RG GTF +G KGGVH+ K + + MLGQ LVTKQT G +V KV + E L + Sbjct: 82 RGLGTFKSGLKGGVHIVKR-DEAEEIAGKMLGQVLVTKQTGPQGKVVSKVYLCEKLSLVN 140 Query: 403 ETYLAILMDRTSGGLVVVSSPQGGMDIEEVAIKNPAAILKTKVDINKGITKSQCDEIAQH 582 E Y +I++DR S G ++++ +GG IE++A K P I+K +D+ GIT ++ Sbjct: 141 EMYFSIILDRKSAGPLIIACKKGGTSIEDLAEKFPDMIIKVPIDVFAGITDEDAAKVVDG 200 Query: 583 LLFTSKQQIDEVGKQLRGLFDLFLQHDCTQIEINPFGVTPGGQIVCFDAKLNFDDNAAFR 762 L + + D + +Q++ L++LF + DCT +EINP T Q+V DAKLNFDDNAAFR Sbjct: 201 LAPKAADRKDSI-EQVKKLYELFRKTDCTMLEINPLAETSTNQLVAADAKLNFDDNAAFR 259 Query: 763 QREIFSLHDPVAEEQDpreIEAAQNGLNYIGLHGNIGCVVNGAGLAMATMDIVKLNGGTP 942 Q+E+F++ DP E Dpre+ AA+ LNYIGL G IGC+VNGAGLAMATMDI+KL+GGTP Sbjct: 260 QKEVFAMRDPTQE--DpreVAAAKVDLNYIGLDGEIGCMVNGAGLAMATMDIIKLHGGTP 317 Query: 943 ANFLDLGGGVTASQVKQAFHILTGDPQVKSIFINIFGGIVNCGTIAEGLTAACRDVELKV 1122 ANFLD+GG + QV +AF ILT D +VK+I +NIFGGI+ C IA G+ A ++V LKV Sbjct: 318 ANFLDVGGNASEHQVVEAFKILTSDDKVKAILVNIFGGIMKCDVIASGIVNAAKEVALKV 377 Query: 1123 PLIVRLEGTNVDSARRILSNSGIPMKMSTDLDEAAKMAVHSVA 1251 P++VRLEGTNV+ +RIL SG+ + + DLD+AA+ AV ++A Sbjct: 378 PVVVRLEGTNVEQGKRILKESGMKLITADDLDDAAEKAVKALA 420
>sp|Q9P2R7|SUCB1_HUMAN Succinyl-CoA ligase [ADP-forming] beta-chain, mitochondrial precursor (Succinyl-CoA synthetase, betaA chain) (SCS-betaA) (ATP-specific succinyl-CoA synthetase beta subunit) Length = 463 Score = 377 bits (967), Expect = e-104 Identities = 191/397 (48%), Positives = 274/397 (69%) Frame = +1 Query: 49 KQTRNLNLQEYQSKGLMSKFGVNIQKFKVASTEQDAIDISKTFKVKEYVIKAQILAGGRG 228 +Q RNL+L EY S L+ + GV++ K VA + +A I+K K+ VIKAQ+LAGGRG Sbjct: 48 QQQRNLSLHEYMSMELLQEAGVSVPKGYVAKSPDEAYAIAKKLGSKDVVIKAQVLAGGRG 107 Query: 229 KGTFSNGFKGGVHLTKNPSDVPNLVKSMLGQRLVTKQTSKDGVMVHKVMVAEALDIDRET 408 KGTF +G KGGV + +P + + M+G++L TKQT + G + ++V+V E RE Sbjct: 108 KGTFESGLKGGVKIVFSPEEAKAVSSQMIGKKLFTKQTGEKGRICNQVLVCERKYPRREY 167 Query: 409 YLAILMDRTSGGLVVVSSPQGGMDIEEVAIKNPAAILKTKVDINKGITKSQCDEIAQHLL 588 Y AI M+R+ G V++ S GG++IE+VA + P AI+K +DI +GI K Q ++AQ + Sbjct: 168 YFAITMERSFQGPVLIGSSHGGVNIEDVAAETPEAIIKEPIDIEEGIKKEQALQLAQKMG 227 Query: 589 FTSKQQIDEVGKQLRGLFDLFLQHDCTQIEINPFGVTPGGQIVCFDAKLNFDDNAAFRQR 768 F ++ + + L+ LFL++D T IEINP G ++C DAK+NFD N+A+RQ+ Sbjct: 228 FPPNI-VESAAENMVKLYSLFLKYDATMIEINPMVEDSDGAVLCMDAKINFDSNSAYRQK 286 Query: 769 EIFSLHDPVAEEQDpreIEAAQNGLNYIGLHGNIGCVVNGAGLAMATMDIVKLNGGTPAN 948 +IF L D E D R+ +AA+ LNYIGL GNIGC+VNGAGLAMATMDI+KL+GGTPAN Sbjct: 287 KIFDLQDWTQE--DERDKDAAKANLNYIGLDGNIGCLVNGAGLAMATMDIIKLHGGTPAN 344 Query: 949 FLDLGGGVTASQVKQAFHILTGDPQVKSIFINIFGGIVNCGTIAEGLTAACRDVELKVPL 1128 FLD+GGG T QV +AF ++T D +V +I +NIFGGI+ C IA+G+ A +D+E+K+P+ Sbjct: 345 FLDVGGGATVHQVTEAFKLITSDKKVLAILVNIFGGIMRCDVIAQGIVMAVKDLEIKIPV 404 Query: 1129 IVRLEGTNVDSARRILSNSGIPMKMSTDLDEAAKMAV 1239 +VRL+GT VD A+ ++++SG+ + DLDEAA+M V Sbjct: 405 VVRLQGTRVDDAKALIADSGLKILACDDLDEAARMVV 441
>sp|P53587|SUCB_NEOFR Succinyl-CoA ligase [GDP-forming] beta-chain, hydrogenosomal precursor (Succinyl-CoA synthetase, beta chain) (SCS-beta) Length = 437 Score = 354 bits (908), Expect = 4e-97 Identities = 187/406 (46%), Positives = 258/406 (63%), Gaps = 1/406 (0%) Frame = +1 Query: 25 SLLSFPLWKQTRNLNLQEYQSKGLMSKFGVNIQKFKVASTEQDAIDISKTFKVKEYVIKA 204 +L S Q R L++ EY S L+ ++ VN K VA T ++A +K ++ VIKA Sbjct: 15 ALASIAQTAQKRFLSVHEYCSMNLLHEYNVNAPKGIVAKTPEEAYQAAKKLNTEDLVIKA 74 Query: 205 QILAGGRGKGTFSNGFKGGVHLTKNPSDVPNLVKSMLGQRLVTKQTSKDGVMVHKVMVAE 384 Q+LAGGRGKG F +G +GGV L P V + MLG +L+TKQT G + V V E Sbjct: 75 QVLAGGRGKGHFDSGLQGGVKLCYTPEQVKDYASKMLGHKLITKQTGAAGRDCNAVYVVE 134 Query: 385 ALDIDRETYLAILMDRTSGGLVVVSSPQGGMDIEEVAIKNPAAILKTKVDINKGITKSQC 564 RE Y AIL+DR + G V+V+S +GG++IEEVA NP AIL I G+T+ Sbjct: 135 RQYAAREYYFAILLDRATRGPVLVASSEGGVEIEEVAKTNPDAILTVPFSIKTGLTREVA 194 Query: 565 DEIAQHLLFTSKQQIDEVGKQLRGLFDLFLQHDCTQIEINPFGVTPGGQIVCFDAKLNFD 744 + A+ + FT K + + L+ +F++ D T +EINP G++VC DAK FD Sbjct: 195 LDTAKKMGFTEKC-VPQAADTFMKLYKIFIEKDATMVEINPMAENNRGEVVCMDAKFGFD 253 Query: 745 DNAAFRQREIFSLHDPVAEEQDpreIEA-AQNGLNYIGLHGNIGCVVNGAGLAMATMDIV 921 DNA+++ + IF+L D E Dpre+ A+ LNY+G+ GNIGC+VNGAGLAMATMDI+ Sbjct: 254 DNASYKPKGIFALRDTTQE--DpreVAGHAKWNLNYVGMEGNIGCLVNGAGLAMATMDII 311 Query: 922 KLNGGTPANFLDLGGGVTASQVKQAFHILTGDPQVKSIFINIFGGIVNCGTIAEGLTAAC 1101 KLNGG PANFLD+GG TA QVK+AF I++ D V +I +NIFGGI+ C +AEG+ A Sbjct: 312 KLNGGVPANFLDVGGSATAKQVKEAFKIISSDKAVSAILVNIFGGIMRCDIVAEGVIQAV 371 Query: 1102 RDVELKVPLIVRLEGTNVDSARRILSNSGIPMKMSTDLDEAAKMAV 1239 +++ L +PL+VRL+GT V+ AR ++ NS + + DLD+AAK V Sbjct: 372 KELGLDIPLVVRLQGTKVEEARELIKNSNLSLYAIDDLDKAAKKVV 417
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 167,370,645
Number of Sequences: 369166
Number of extensions: 3713271
Number of successful extensions: 10695
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 9802
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10440
length of database: 68,354,980
effective HSP length: 114
effective length of database: 47,295,190
effective search space used: 16647906880
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail