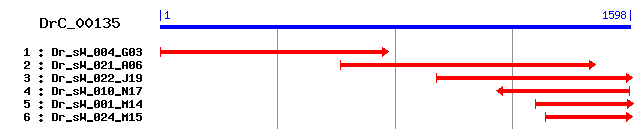
DrC_00135
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00135 (1598 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|O08762|NETR_MOUSE Neurotrypsin precursor (Motopsin) (Bra... 147 9e-35 sp|Q5G265|NETR_SAGLB Neurotrypsin precursor 147 9e-35 sp|Q5G269|NETR_PONPY Neurotrypsin precursor 142 3e-33 sp|P56730|NETR_HUMAN Neurotrypsin precursor (Motopsin) (Ley... 142 4e-33 sp|Q5G271|NETR_PANTR Neurotrypsin precursor 142 4e-33 sp|Q5G268|NETR_HYLLE Neurotrypsin precursor 141 5e-33 sp|Q5G270|NETR_GORGO Neurotrypsin precursor 141 6e-33 sp|Q5G267|NETR_MACMU Neurotrypsin precursor 140 8e-33 sp|Q5G266|NETR_TRAPH Neurotrypsin precursor 140 1e-32 sp|Q9BYE2|TMPSD_HUMAN Transmembrane protease, serine 13 (Mo... 99 3e-20
>sp|O08762|NETR_MOUSE Neurotrypsin precursor (Motopsin) (Brain-specific serine protease 3) (BSSP-3) Length = 761 Score = 147 bits (371), Expect = 9e-35 Identities = 130/477 (27%), Positives = 191/477 (40%), Gaps = 45/477 (9%) Frame = +2 Query: 200 GHFQGYMLYNDSMICMDSFAQNNAEVYCRYLGYP-GARKFXXXXXXXXXXXXLFNLQCNG 376 G + Y +C D + + N V CR LG+ G + L ++ C+G Sbjct: 284 GRLEVYYKGQWGTVCDDGWTEMNTYVACRLLGFKYGKQSSVNHFDGSNRPIWLDDVSCSG 343 Query: 377 NENSPAEC--RVYTNSYCTSGRALGTVCQT------------VALAGTNRSNEGRIEVYN 514 E S +C R + C+ +G C + L EGR+EV+ Sbjct: 344 KEVSFIQCSRRQWGRHDCSHREDVGLTCYPDSDGHRLSPGFPIRLVDGENKKEGRVEVFV 403 Query: 515 AGQKGLMCDDTFNADIANIVCRQLGFSFALNQTTAAGYVYGFGNKNFLLDYLYCNGNEDS 694 GQ G +CDD + A ++CRQLG+ T A +G G +D + C GNE + Sbjct: 404 NGQWGTICDDGWTDKHAAVICRQLGYKGPARARTMA--YFGEGKGPIHMDNVKCTGNEKA 461 Query: 695 IDDCPWQYWRLRNCYGNDVAGVYC---------AGN-----ASCGEVYTEERNQLSSFGY 832 + DC Q NC ++ AGV C +GN + CG R + G Sbjct: 462 LADCVKQDIGRHNCRHSEDAGVICDYLEKKASSSGNKEMLSSGCGLRLLHRRQKRIIGGN 521 Query: 833 NTTFGANPWQVRIIPRVANE----WCSGTIINKNWVLTSANCFTMSYDNFXXXXXXXXXX 1000 N+ GA PWQ + R A+ C T+++ WVLT+A+CF +N Sbjct: 522 NSLRGAWPWQASLRLRSAHGDGRLLCGATLLSSCWVLTAAHCFKRYGNNSRSYAVRVGDY 581 Query: 1001 XXXXNWAYTNNI--NSVFIHPQYSAVTGDYNLALVKTSP-----VAFDINVQPACLPSKN 1159 + I + IH Y DY++ALV+ +V PACLP Sbjct: 582 HTLVPEEFEQEIGVQQIVIHRNYRPDRSDYDIALVRLQGPGEQCARLSTHVLPACLPLWR 641 Query: 1160 LVP--NSLDCITAGWNHKLETNSRPLLGKSVNLYNTTNCKSLTQFDDDKLARYFCSDNNM 1333 P + +C GW SR L +V L CK ++ R C+ N Sbjct: 642 ERPQKTASNCHITGWGDTGRAYSRTLQQAAVPLLPKRFCKE--RYKGLFTGRMLCAGNLQ 699 Query: 1334 GSSAI--FNGESGGPLIC-SIDGSKVIAGVVSKSINSARATSIRLFTRVDYMLPWIK 1495 + + G+SGGPL+C D S V+ GV S + ++TRV +PWIK Sbjct: 700 EDNRVDSCQGDSGGPLMCEKPDESWVVYGVTSWGYGCGVKDTPGVYTRVPAFVPWIK 756
Score = 87.0 bits (214), Expect = 1e-16
Identities = 47/116 (40%), Positives = 64/116 (55%)
Frame = +2
Query: 419 YCTSGRALGTVCQTVALAGTNRSNEGRIEVYNAGQKGLMCDDTFNADIANIVCRQLGFSF 598
YC G+ G + L G N +EGR+E+Y+AGQ G +CDD ++ A+++CRQLG S
Sbjct: 154 YCDCGQ--GPALPVIRLVGGNSGHEGRVELYHAGQWGTICDDQWDNADADVICRQLGLSG 211
Query: 599 ALNQTTAAGYVYGFGNKNFLLDYLYCNGNEDSIDDCPWQYWRLRNCYGNDVAGVYC 766
A +G G+ LLD + C GNE SI+ CP W NC + AGV C
Sbjct: 212 IAKAWHQAH--FGEGSGPILLDEVRCTGNELSIEQCPKSSWGEHNCGHKEDAGVSC 265
Score = 80.1 bits (196), Expect = 2e-14
Identities = 65/239 (27%), Positives = 101/239 (42%), Gaps = 10/239 (4%)
Frame = +2
Query: 239 ICMDSFAQNNAEVYCRYLGYPGARKFXXXXXXXXXXXXLF--NLQCNGNENSPAEC--RV 406
IC D + +A+V CR LG G K + ++C GNE S +C
Sbjct: 190 ICDDQWDNADADVICRQLGLSGIAKAWHQAHFGEGSGPILLDEVRCTGNELSIEQCPKSS 249
Query: 407 YTNSYCTSGRALGTVCQ-----TVALAGTNRSNEGRIEVYNAGQKGLMCDDTFNADIANI 571
+ C G C + LAG ++EGR+EVY GQ G +CDD + +
Sbjct: 250 WGEHNCGHKEDAGVSCVPLTDGVIRLAGGKSTHEGRLEVYYKGQWGTVCDDGWTEMNTYV 309
Query: 572 VCRQLGFSFALNQTTAAGYVYGFGNKNFLLDYLYCNGNEDSIDDCPWQYWRLRNCYGNDV 751
CR LGF + + ++ + G N+ LD + C+G E S C + W +C +
Sbjct: 310 ACRLLGFKY--GKQSSVNHFDG-SNRPIWLDDVSCSGKEVSFIQCSRRQWGRHDCSHRED 366
Query: 752 AGVYCAGNASCGEVYTEERNQLS-SFGYNTTFGANPWQVRIIPRVANEWCSGTIINKNW 925
G+ C ++ + ++LS F G N + R+ V +W GTI + W
Sbjct: 367 VGLTCYPDS--------DGHRLSPGFPIRLVDGENKKEGRVEVFVNGQW--GTICDDGW 415
Score = 37.4 bits (85), Expect = 0.12
Identities = 24/104 (23%), Positives = 41/104 (39%), Gaps = 4/104 (3%)
Frame = +2
Query: 194 KPGHFQGYMLYNDSMICMDSFAQNNAEVYCRYLGY--PGARKFXXXXXXXXXXXXLFNLQ 367
K G + ++ IC D + +A V CR LGY P + + N++
Sbjct: 395 KEGRVEVFVNGQWGTICDDGWTDKHAAVICRQLGYKGPARARTMAYFGEGKGPIHMDNVK 454
Query: 368 CNGNENSPAEC--RVYTNSYCTSGRALGTVCQTVALAGTNRSNE 493
C GNE + A+C + C G +C + ++ N+
Sbjct: 455 CTGNEKALADCVKQDIGRHNCRHSEDAGVICDYLEKKASSSGNK 498
>sp|Q5G265|NETR_SAGLB Neurotrypsin precursor Length = 875 Score = 147 bits (371), Expect = 9e-35 Identities = 127/477 (26%), Positives = 196/477 (41%), Gaps = 45/477 (9%) Frame = +2 Query: 200 GHFQGYMLYNDSMICMDSFAQNNAEVYCRYLGYP-GARKFXXXXXXXXXXXXLFNLQCNG 376 G + Y +C D + + N V CR LG+ G + L ++ C+G Sbjct: 398 GRLEVYYSGQWGTVCDDGWTELNTYVVCRQLGFKYGKQASANHFEESAGPIWLDDVSCSG 457 Query: 377 NENSPAEC--RVYTNSYCTSGRALGTVCQT------------VALAGTNRSNEGRIEVYN 514 E +C R + C+ +G C V L EGR+EV+ Sbjct: 458 KETRFLQCSRRQWGRHDCSHREDVGIACYPGSEGHRLSLGFPVRLMDGENKKEGRVEVFI 517 Query: 515 AGQKGLMCDDTFNADIANIVCRQLGFSFALNQTTAAGYVYGFGNKNFLLDYLYCNGNEDS 694 GQ G +CDD + A ++CRQLG+ T A +G G +D + C GNE S Sbjct: 518 NGQWGTICDDGWTDKDAAVICRQLGYKGPARARTMA--YFGEGKGPIHVDNVKCTGNERS 575 Query: 695 IDDCPWQYWRLRNCYGNDVAGVYC-------AGNAS-------CGEVYTEERNQLSSFGY 832 + DC Q NC ++ AGV C +GN++ CG R + G Sbjct: 576 LADCIKQDIGRHNCRHSEDAGVICDYFGKKASGNSNKESLSSVCGLRLLHRRQKRIIGGK 635 Query: 833 NTTFGANPWQVRIIPRVANE----WCSGTIINKNWVLTSANCFTMSYDNFXXXXXXXXXX 1000 N+ G PWQV + + ++ C T+++ WVLT+A+CF ++ Sbjct: 636 NSLRGGWPWQVSLRLKSSHRDGRLLCGATLLSSCWVLTAAHCFKRYGNSTRNYAVRVGDY 695 Query: 1001 XXXXNWAYTNNI--NSVFIHPQYSAVTGDYNLALVKTSP-----VAFDINVQPACLPSKN 1159 + I + IH +Y + DY++ALV+ F+ +V PACLP Sbjct: 696 HTLVPEEFEEEIGVQEIVIHREYRPDSSDYDIALVRLQGPEEQCARFNSHVLPACLPLWR 755 Query: 1160 LVP--NSLDCITAGWNHKLETNSRPLLGKSVNLYNTTNCKSLTQFDDDKLARYFCSDNNM 1333 P + +C GW + SR L +++L C+ ++ R C+ N Sbjct: 756 ERPQKTASNCYITGWGDTGQAYSRTLQQAAIHLLPKRFCEE--RYKGRFTGRMLCAGNLH 813 Query: 1334 GSSAI--FNGESGGPLICSIDG-SKVIAGVVSKSINSARATSIRLFTRVDYMLPWIK 1495 + G+SGGPL+C G S V+ GV S S ++T+V +PWIK Sbjct: 814 EHKHVDSCQGDSGGPLMCERPGESWVVYGVTSWGYGCGVKDSPGVYTKVSAFVPWIK 870
Score = 89.0 bits (219), Expect = 4e-17
Identities = 65/228 (28%), Positives = 100/228 (43%), Gaps = 12/228 (5%)
Frame = +2
Query: 119 INTNYFYCYSGSLQLKCQDKLQFTLKPGHFQGYMLYNDSMICMDSFAQNNAEVYCRYL-- 292
++ Y C GS++L+ K +F G + Y +C + ++A V C L
Sbjct: 158 VDWGYCDCRHGSVRLR-GSKNEFE---GTVEVYANGVWGTVCSSHWDDSDASVICHQLQL 213
Query: 293 GYPGARKFXXXXXXXXXXXXLFNLQCNGNENSPAECR--VYTNSYCTSGRALGTVCQ--- 457
G G K N++C G+E + C ++ C A +C
Sbjct: 214 GGKGIAKQTPFSGLGLIPVYWSNVRCRGDEENILLCEKDIWQGGVCPQKMAAAVMCSFSH 273
Query: 458 -----TVALAGTNRSNEGRIEVYNAGQKGLMCDDTFNADIANIVCRQLGFSFALNQTTAA 622
+ L G + +EGR+E+Y+AGQ G +CDD ++ A ++CRQL S A
Sbjct: 274 GPAFPIIRLVGGSSVHEGRVELYHAGQWGTICDDQWDDADAEVICRQLSLSGIAKAWHQA 333
Query: 623 GYVYGFGNKNFLLDYLYCNGNEDSIDDCPWQYWRLRNCYGNDVAGVYC 766
+G G+ +LD + C GNE SI+ CP W NC + AGV C
Sbjct: 334 --YFGEGSGPVMLDEVRCTGNELSIEQCPKSSWGEHNCGHKEDAGVSC 379
Score = 82.0 bits (201), Expect = 4e-15
Identities = 69/239 (28%), Positives = 99/239 (41%), Gaps = 10/239 (4%)
Frame = +2
Query: 239 ICMDSFAQNNAEVYCRYLGYPGARK--FXXXXXXXXXXXXLFNLQCNGNENSPAEC--RV 406
IC D + +AEV CR L G K L ++C GNE S +C
Sbjct: 304 ICDDQWDDADAEVICRQLSLSGIAKAWHQAYFGEGSGPVMLDEVRCTGNELSIEQCPKSS 363
Query: 407 YTNSYCTSGRALGTVCQ-----TVALAGTNRSNEGRIEVYNAGQKGLMCDDTFNADIANI 571
+ C G C + LAG S+EGR+EVY +GQ G +CDD + +
Sbjct: 364 WGEHNCGHKEDAGVSCTPLTDGVIRLAGGKGSHEGRLEVYYSGQWGTVCDDGWTELNTYV 423
Query: 572 VCRQLGFSFALNQTTAAGYVYGFGNKNFLLDYLYCNGNEDSIDDCPWQYWRLRNCYGNDV 751
VCRQLGF + Q +A + G LD + C+G E C + W +C +
Sbjct: 424 VCRQLGFKYG-KQASANHFEESAG--PIWLDDVSCSGKETRFLQCSRRQWGRHDCSHRED 480
Query: 752 AGVYCAGNASCGEVYTEERNQLS-SFGYNTTFGANPWQVRIIPRVANEWCSGTIINKNW 925
G+ C + E ++LS F G N + R+ + +W GTI + W
Sbjct: 481 VGIACYPGS--------EGHRLSLGFPVRLMDGENKKEGRVEVFINGQW--GTICDDGW 529
Score = 42.4 bits (98), Expect = 0.004
Identities = 36/127 (28%), Positives = 54/127 (42%), Gaps = 10/127 (7%)
Frame = +2
Query: 419 YCTSGRALGTV------CQ--TVALAGTNRSNEGRIEVYNAGQKGLMCDDTFNADIANIV 574
+C G A G V C+ +V L G+ EG +EVY G G +C ++ A+++
Sbjct: 148 WCFYGDARGKVDWGYCDCRHGSVRLRGSKNEFEGTVEVYANGVWGTVCSSHWDDSDASVI 207
Query: 575 CRQL--GFSFALNQTTAAGYVYGFGNKNFLLDYLYCNGNEDSIDDCPWQYWRLRNCYGND 748
C QL G QT +G G + C G+E++I C W+ C
Sbjct: 208 CHQLQLGGKGIAKQTPFSG----LGLIPVYWSNVRCRGDEENILLCEKDIWQGGVCPQKM 263
Query: 749 VAGVYCA 769
A V C+
Sbjct: 264 AAAVMCS 270
Score = 39.7 bits (91), Expect = 0.025
Identities = 26/104 (25%), Positives = 40/104 (38%), Gaps = 4/104 (3%)
Frame = +2
Query: 194 KPGHFQGYMLYNDSMICMDSFAQNNAEVYCRYLGYPGARKFXXXXXXXXXXXXLF--NLQ 367
K G + ++ IC D + +A V CR LGY G + + N++
Sbjct: 509 KEGRVEVFINGQWGTICDDGWTDKDAAVICRQLGYKGPARARTMAYFGEGKGPIHVDNVK 568
Query: 368 CNGNENSPAEC--RVYTNSYCTSGRALGTVCQTVALAGTNRSNE 493
C GNE S A+C + C G +C + SN+
Sbjct: 569 CTGNERSLADCIKQDIGRHNCRHSEDAGVICDYFGKKASGNSNK 612
>sp|Q5G269|NETR_PONPY Neurotrypsin precursor Length = 877 Score = 142 bits (358), Expect = 3e-33 Identities = 127/478 (26%), Positives = 192/478 (40%), Gaps = 46/478 (9%) Frame = +2 Query: 200 GHFQGYMLYNDSMICMDSFAQNNAEVYCRYLGYP-GARKFXXXXXXXXXXXXLFNLQCNG 376 G + Y +C D + + N V CR LG+ G + L ++ C+G Sbjct: 400 GRLEVYYRGQWGTVCDDGWTELNTYVVCRQLGFKFGKQASANHFEESTGPIWLDDVSCSG 459 Query: 377 NENSPAEC--RVYTNSYCTSGRALGTVCQT------------VALAGTNRSNEGRIEVYN 514 E +C R + C+ + C V L EGR+EV+ Sbjct: 460 KETRFLQCSRRQWGRHDCSHREDVSIACYPGGEGHRLSLGFPVRLMDGENKKEGRVEVFI 519 Query: 515 AGQKGLMCDDTFNADIANIVCRQLGFSFALNQTTAAGYVYGFGNKNFLLDYLYCNGNEDS 694 GQ G +CDD + A ++CRQLG+ T A +G G +D + C GNE S Sbjct: 520 NGQWGTICDDGWTDKDAAVICRQLGYKGPARARTMA--YFGEGKGPIHVDNVRCTGNERS 577 Query: 695 IDDCPWQYWRLRNCYGNDVAGVYC-------AGNAS-------CGEVYTEERNQLSSFGY 832 + DC Q NC ++ AGV C +GN++ CG R + G Sbjct: 578 LADCIKQDIGRHNCRHSEDAGVICDYFGKKASGNSNKESLSSVCGLRLLHRRQKRIIGGK 637 Query: 833 NTTFGANPWQVRIIPRVANE----WCSGTIINKNWVLTSANCFTMSYDNF---XXXXXXX 991 N+ G PWQV + + ++ C T+++ WVLT+A+CF Y N Sbjct: 638 NSLRGGWPWQVSLRLKSSHGDGRLLCGATLLSSCWVLTAAHCF-KRYGNSTRNYAVRVGD 696 Query: 992 XXXXXXXNWAYTNNINSVFIHPQYSAVTGDYNLALVKTSP-----VAFDINVQPACLPSK 1156 + + + IH +Y + DY++ALV+ F +V PACLP Sbjct: 697 YHTLVPEEFEEEVGVQQIVIHREYRPDSSDYDIALVRLQGPEEQCARFSSHVLPACLPLW 756 Query: 1157 NLVP--NSLDCITAGWNHKLETNSRPLLGKSVNLYNTTNCKSLTQFDDDKLARYFCSDNN 1330 P + +C GW SR L ++ L C+ ++ R C+ N Sbjct: 757 RERPQKTASNCYITGWGDTGRAYSRTLQQAAIPLLPKRFCEE--RYKGXFTGRMLCAGNL 814 Query: 1331 MGSSAI--FNGESGGPLICSIDG-SKVIAGVVSKSINSARATSIRLFTRVDYMLPWIK 1495 + G+SGGPL+C G S V+ GV S S ++T+V +PWIK Sbjct: 815 HEHKRVDSCQGDSGGPLMCERPGESWVVYGVTSWGYGCGVKDSPGVYTKVSAFVPWIK 872
Score = 88.6 bits (218), Expect = 5e-17
Identities = 65/228 (28%), Positives = 99/228 (43%), Gaps = 12/228 (5%)
Frame = +2
Query: 119 INTNYFYCYSGSLQLKCQDKLQFTLKPGHFQGYMLYNDSMICMDSFAQNNAEVYCRYL-- 292
++ Y C GS++L+ K +F G + Y +C + ++A V C L
Sbjct: 160 VDWGYCDCRHGSVRLR-GGKNEFE---GTVEVYASGVWGTVCSSHWDDSDASVICHQLQL 215
Query: 293 GYPGARKFXXXXXXXXXXXXLFNLQCNGNENSPAECR--VYTNSYCTSGRALGTVCQ--- 457
G G K N++C G+E + C ++ C A C
Sbjct: 216 GGKGIAKQTPFSGLGLIPIYWSNVRCQGDEENILLCEKDIWQGGVCPQKMAAAVTCSFSH 275
Query: 458 -----TVALAGTNRSNEGRIEVYNAGQKGLMCDDTFNADIANIVCRQLGFSFALNQTTAA 622
+ L G + +EGR+E+Y+AGQ G +CDD ++ A ++CRQL S A
Sbjct: 276 GPTFPIIRLVGGSSVHEGRVELYHAGQWGTVCDDQWDDADAEVICRQLSLSGIAKAWHQA 335
Query: 623 GYVYGFGNKNFLLDYLYCNGNEDSIDDCPWQYWRLRNCYGNDVAGVYC 766
+G G+ +LD + C GNE SI+ CP W NC + AGV C
Sbjct: 336 --YFGEGSGPVMLDEVRCTGNELSIEQCPKSSWGEHNCGHKEDAGVSC 381
Score = 79.3 bits (194), Expect = 3e-14
Identities = 68/239 (28%), Positives = 96/239 (40%), Gaps = 10/239 (4%)
Frame = +2
Query: 239 ICMDSFAQNNAEVYCRYLGYPGARK--FXXXXXXXXXXXXLFNLQCNGNENSPAEC--RV 406
+C D + +AEV CR L G K L ++C GNE S +C
Sbjct: 306 VCDDQWDDADAEVICRQLSLSGIAKAWHQAYFGEGSGPVMLDEVRCTGNELSIEQCPKSS 365
Query: 407 YTNSYCTSGRALGTVCQ-----TVALAGTNRSNEGRIEVYNAGQKGLMCDDTFNADIANI 571
+ C G C + LAG S+EGR+EVY GQ G +CDD + +
Sbjct: 366 WGEHNCGHKEDAGVSCTPLTDGVIRLAGGKGSHEGRLEVYYRGQWGTVCDDGWTELNTYV 425
Query: 572 VCRQLGFSFALNQTTAAGYVYGFGNKNFLLDYLYCNGNEDSIDDCPWQYWRLRNCYGNDV 751
VCRQLGF F Q +A + G LD + C+G E C + W +C +
Sbjct: 426 VCRQLGFKFG-KQASANHFEESTG--PIWLDDVSCSGKETRFLQCSRRQWGRHDCSHRED 482
Query: 752 AGVYCAGNASCGEVYTEERNQLS-SFGYNTTFGANPWQVRIIPRVANEWCSGTIINKNW 925
+ C E ++LS F G N + R+ + +W GTI + W
Sbjct: 483 VSIACYPGG--------EGHRLSLGFPVRLMDGENKKEGRVEVFINGQW--GTICDDGW 531
Score = 39.7 bits (91), Expect = 0.025
Identities = 26/104 (25%), Positives = 40/104 (38%), Gaps = 4/104 (3%)
Frame = +2
Query: 194 KPGHFQGYMLYNDSMICMDSFAQNNAEVYCRYLGYPGARKFXXXXXXXXXXXXLF--NLQ 367
K G + ++ IC D + +A V CR LGY G + + N++
Sbjct: 511 KEGRVEVFINGQWGTICDDGWTDKDAAVICRQLGYKGPARARTMAYFGEGKGPIHVDNVR 570
Query: 368 CNGNENSPAEC--RVYTNSYCTSGRALGTVCQTVALAGTNRSNE 493
C GNE S A+C + C G +C + SN+
Sbjct: 571 CTGNERSLADCIKQDIGRHNCRHSEDAGVICDYFGKKASGNSNK 614
>sp|P56730|NETR_HUMAN Neurotrypsin precursor (Motopsin) (Leydin) Length = 875 Score = 142 bits (357), Expect = 4e-33 Identities = 128/481 (26%), Positives = 192/481 (39%), Gaps = 49/481 (10%) Frame = +2 Query: 200 GHFQGYMLYNDSMICMDSFAQNNAEVYCRYLGYP-GARKFXXXXXXXXXXXXLFNLQCNG 376 G + Y +C D + + N V CR LG+ G + L ++ C+G Sbjct: 398 GRLEVYYRGQWGTVCDDGWTELNTYVVCRQLGFKYGKQASANHFEESTGPIWLDDVSCSG 457 Query: 377 NENSPAEC--RVYTNSYCTSGRALGTVCQT------------VALAGTNRSNEGRIEVYN 514 E +C R + C+ + C V L EGR+EV+ Sbjct: 458 KETRFLQCSRRQWGRHDCSHREDVSIACYPGGEGHRLSLGFPVRLMDGENKKEGRVEVFI 517 Query: 515 AGQKGLMCDDTFNADIANIVCRQLGFSFALNQTTAAGYVYGFGNKNFLLDYLYCNGNEDS 694 GQ G +CDD + A ++CRQLG+ T A +G G +D + C GNE S Sbjct: 518 NGQWGTICDDGWTDKDAAVICRQLGYKGPARARTMA--YFGEGKGPIHVDNVKCTGNERS 575 Query: 695 IDDCPWQYWRLRNCYGNDVAGVYC-------AGNAS-------CGEVYTEERNQLSSFGY 832 + DC Q NC ++ AGV C +GN++ CG R + G Sbjct: 576 LADCIKQDIGRHNCRHSEDAGVICDYFGKKASGNSNKESLSSVCGLRLLHRRQKRIIGGK 635 Query: 833 NTTFGANPWQVRIIPRVANE----WCSGTIINKNWVLTSANCF------TMSYDNFXXXX 982 N+ G PWQV + + ++ C T+++ WVLT+A+CF T SY Sbjct: 636 NSLRGGWPWQVSLRLKSSHGDGRLLCGATLLSSCWVLTAAHCFKRYGNSTRSY----AVR 691 Query: 983 XXXXXXXXXXNWAYTNNINSVFIHPQYSAVTGDYNLALVKTSP-----VAFDINVQPACL 1147 + + + IH +Y DY++ALV+ F +V PACL Sbjct: 692 VGDYHTLVPEEFEEEIGVQQIVIHREYRPDRSDYDIALVRLQGPEEQCARFSSHVLPACL 751 Query: 1148 PSKNLVP--NSLDCITAGWNHKLETNSRPLLGKSVNLYNTTNCKSLTQFDDDKLARYFCS 1321 P P + +C GW SR L ++ L C+ ++ R C+ Sbjct: 752 PLWRERPQKTASNCYITGWGDTGRAYSRTLQQAAIPLLPKRFCEE--RYKGRFTGRMLCA 809 Query: 1322 DNNMGSSAI--FNGESGGPLICSIDG-SKVIAGVVSKSINSARATSIRLFTRVDYMLPWI 1492 N + G+SGGPL+C G S V+ GV S S ++T+V +PWI Sbjct: 810 GNLHEHKRVDSCQGDSGGPLMCERPGESWVVYGVTSWGYGCGVKDSPGVYTKVSAFVPWI 869 Query: 1493 K 1495 K Sbjct: 870 K 870
Score = 92.4 bits (228), Expect = 3e-18
Identities = 67/228 (29%), Positives = 101/228 (44%), Gaps = 12/228 (5%)
Frame = +2
Query: 119 INTNYFYCYSGSLQLKCQDKLQFTLKPGHFQGYMLYNDSMICMDSFAQNNAEVYCRYL-- 292
++ Y C GS++L+ K +F G + Y +C + ++A V C L
Sbjct: 158 VDWGYCDCRHGSVRLR-GGKNEFE---GTVEVYASGVWGTVCSSHWDDSDASVICHQLQL 213
Query: 293 GYPGARKFXXXXXXXXXXXXLFNLQCNGNENSPAECR--VYTNSYCTSGRALGTVCQ--- 457
G G K N++C G+E + C ++ C A C
Sbjct: 214 GGKGIAKQTPFSGLGLIPIYWSNVRCRGDEENILLCEKDIWQGGVCPQKMAAAVTCSFSH 273
Query: 458 -----TVALAGTNRSNEGRIEVYNAGQKGLMCDDTFNADIANIVCRQLGFSFALNQTTAA 622
+ LAG + +EGR+E+Y+AGQ G +CDD ++ A ++CRQLG S A
Sbjct: 274 GPTFPIIRLAGGSSVHEGRVELYHAGQWGTVCDDQWDDADAEVICRQLGLSGIAKAWHQA 333
Query: 623 GYVYGFGNKNFLLDYLYCNGNEDSIDDCPWQYWRLRNCYGNDVAGVYC 766
+G G+ +LD + C GNE SI+ CP W NC + AGV C
Sbjct: 334 --YFGEGSGPVMLDEVRCTGNELSIEQCPKSSWGEHNCGHKEDAGVSC 379
Score = 80.5 bits (197), Expect = 1e-14
Identities = 68/239 (28%), Positives = 97/239 (40%), Gaps = 10/239 (4%)
Frame = +2
Query: 239 ICMDSFAQNNAEVYCRYLGYPGARK--FXXXXXXXXXXXXLFNLQCNGNENSPAEC--RV 406
+C D + +AEV CR LG G K L ++C GNE S +C
Sbjct: 304 VCDDQWDDADAEVICRQLGLSGIAKAWHQAYFGEGSGPVMLDEVRCTGNELSIEQCPKSS 363
Query: 407 YTNSYCTSGRALGTVCQ-----TVALAGTNRSNEGRIEVYNAGQKGLMCDDTFNADIANI 571
+ C G C + LAG S+EGR+EVY GQ G +CDD + +
Sbjct: 364 WGEHNCGHKEDAGVSCTPLTDGVIRLAGGKGSHEGRLEVYYRGQWGTVCDDGWTELNTYV 423
Query: 572 VCRQLGFSFALNQTTAAGYVYGFGNKNFLLDYLYCNGNEDSIDDCPWQYWRLRNCYGNDV 751
VCRQLGF + Q +A + G LD + C+G E C + W +C +
Sbjct: 424 VCRQLGFKYG-KQASANHFEESTG--PIWLDDVSCSGKETRFLQCSRRQWGRHDCSHRED 480
Query: 752 AGVYCAGNASCGEVYTEERNQLS-SFGYNTTFGANPWQVRIIPRVANEWCSGTIINKNW 925
+ C E ++LS F G N + R+ + +W GTI + W
Sbjct: 481 VSIACYPGG--------EGHRLSLGFPVRLMDGENKKEGRVEVFINGQW--GTICDDGW 529
Score = 39.7 bits (91), Expect = 0.025
Identities = 26/104 (25%), Positives = 40/104 (38%), Gaps = 4/104 (3%)
Frame = +2
Query: 194 KPGHFQGYMLYNDSMICMDSFAQNNAEVYCRYLGYPGARKFXXXXXXXXXXXXLF--NLQ 367
K G + ++ IC D + +A V CR LGY G + + N++
Sbjct: 509 KEGRVEVFINGQWGTICDDGWTDKDAAVICRQLGYKGPARARTMAYFGEGKGPIHVDNVK 568
Query: 368 CNGNENSPAEC--RVYTNSYCTSGRALGTVCQTVALAGTNRSNE 493
C GNE S A+C + C G +C + SN+
Sbjct: 569 CTGNERSLADCIKQDIGRHNCRHSEDAGVICDYFGKKASGNSNK 612
>sp|Q5G271|NETR_PANTR Neurotrypsin precursor Length = 875 Score = 142 bits (357), Expect = 4e-33 Identities = 128/481 (26%), Positives = 192/481 (39%), Gaps = 49/481 (10%) Frame = +2 Query: 200 GHFQGYMLYNDSMICMDSFAQNNAEVYCRYLGYP-GARKFXXXXXXXXXXXXLFNLQCNG 376 G + Y +C D + + N V CR LG+ G + L ++ C+G Sbjct: 398 GRLEVYYRGQWGTVCDDGWTELNTYVVCRQLGFKYGKQASANHFEESTGPIWLDDVSCSG 457 Query: 377 NENSPAEC--RVYTNSYCTSGRALGTVCQT------------VALAGTNRSNEGRIEVYN 514 E +C R + C+ + C V L EGR+EV+ Sbjct: 458 KETRFLQCSRRQWGRHDCSHREDVSIACYPGGEGHRLSLGFPVRLVDGENKKEGRVEVFI 517 Query: 515 AGQKGLMCDDTFNADIANIVCRQLGFSFALNQTTAAGYVYGFGNKNFLLDYLYCNGNEDS 694 GQ G +CDD + A ++CRQLG+ T A +G G +D + C GNE S Sbjct: 518 NGQWGTICDDGWTDKDAAVICRQLGYKGPARARTMA--YFGEGKGPIHVDNVKCTGNERS 575 Query: 695 IDDCPWQYWRLRNCYGNDVAGVYC-------AGNAS-------CGEVYTEERNQLSSFGY 832 + DC Q NC ++ AGV C +GN++ CG R + G Sbjct: 576 LADCIKQDIGRHNCRHSEDAGVICDYFGKKASGNSNKESLSSVCGLRLLHRRQKRIIGGK 635 Query: 833 NTTFGANPWQVRIIPRVANE----WCSGTIINKNWVLTSANCF------TMSYDNFXXXX 982 N+ G PWQV + + ++ C T+++ WVLT+A+CF T SY Sbjct: 636 NSLRGGWPWQVSLRLKSSHGDGRLLCGATLLSSCWVLTAAHCFKRYGNSTRSY----AVR 691 Query: 983 XXXXXXXXXXNWAYTNNINSVFIHPQYSAVTGDYNLALVKTSP-----VAFDINVQPACL 1147 + + + IH +Y DY++ALV+ F +V PACL Sbjct: 692 VGDYHTLVPEEFEEEIGVQQIVIHREYRPDRSDYDIALVRLQGPEEQCARFSSHVLPACL 751 Query: 1148 PSKNLVP--NSLDCITAGWNHKLETNSRPLLGKSVNLYNTTNCKSLTQFDDDKLARYFCS 1321 P P + +C GW SR L ++ L C+ ++ R C+ Sbjct: 752 PLWRERPQKTASNCYITGWGDTGRAYSRTLQQAAIPLLPKRFCEE--RYKRRFTGRMLCA 809 Query: 1322 DNNMGSSAI--FNGESGGPLICSIDG-SKVIAGVVSKSINSARATSIRLFTRVDYMLPWI 1492 N + G+SGGPL+C G S V+ GV S S ++T+V +PWI Sbjct: 810 GNLHEHKRVDSCQGDSGGPLMCERPGESWVVYGVTSWGYGCGVKDSPGVYTKVSAFVPWI 869 Query: 1493 K 1495 K Sbjct: 870 K 870
Score = 92.4 bits (228), Expect = 3e-18
Identities = 67/228 (29%), Positives = 101/228 (44%), Gaps = 12/228 (5%)
Frame = +2
Query: 119 INTNYFYCYSGSLQLKCQDKLQFTLKPGHFQGYMLYNDSMICMDSFAQNNAEVYCRYL-- 292
++ Y C GS++L+ K +F G + Y +C + ++A V C L
Sbjct: 158 VDWGYCDCRHGSVRLR-GGKNEFE---GTVEVYASGVWGTVCSSHWDDSDASVICHQLQL 213
Query: 293 GYPGARKFXXXXXXXXXXXXLFNLQCNGNENSPAECR--VYTNSYCTSGRALGTVCQ--- 457
G G K N++C G+E + C ++ C A C
Sbjct: 214 GGKGIAKQTPFSGLGLIPIYWSNVRCRGDEENILLCEKDIWQGGVCPQKMAAAVTCSFSH 273
Query: 458 -----TVALAGTNRSNEGRIEVYNAGQKGLMCDDTFNADIANIVCRQLGFSFALNQTTAA 622
+ LAG + +EGR+E+Y+AGQ G +CDD ++ A ++CRQLG S A
Sbjct: 274 GPTFPIIRLAGGSSVHEGRVELYHAGQWGTVCDDQWDDADAEVICRQLGLSGIAKAWHQA 333
Query: 623 GYVYGFGNKNFLLDYLYCNGNEDSIDDCPWQYWRLRNCYGNDVAGVYC 766
+G G+ +LD + C GNE SI+ CP W NC + AGV C
Sbjct: 334 --YFGEGSGPVMLDEVRCTGNELSIEQCPKSSWGEHNCGHKEDAGVSC 379
Score = 80.9 bits (198), Expect = 1e-14
Identities = 68/239 (28%), Positives = 97/239 (40%), Gaps = 10/239 (4%)
Frame = +2
Query: 239 ICMDSFAQNNAEVYCRYLGYPGARK--FXXXXXXXXXXXXLFNLQCNGNENSPAEC--RV 406
+C D + +AEV CR LG G K L ++C GNE S +C
Sbjct: 304 VCDDQWDDADAEVICRQLGLSGIAKAWHQAYFGEGSGPVMLDEVRCTGNELSIEQCPKSS 363
Query: 407 YTNSYCTSGRALGTVCQ-----TVALAGTNRSNEGRIEVYNAGQKGLMCDDTFNADIANI 571
+ C G C + LAG S+EGR+EVY GQ G +CDD + +
Sbjct: 364 WGEHNCGHKEDAGVSCTPLTDGVIRLAGGKGSHEGRLEVYYRGQWGTVCDDGWTELNTYV 423
Query: 572 VCRQLGFSFALNQTTAAGYVYGFGNKNFLLDYLYCNGNEDSIDDCPWQYWRLRNCYGNDV 751
VCRQLGF + Q +A + G LD + C+G E C + W +C +
Sbjct: 424 VCRQLGFKYG-KQASANHFEESTG--PIWLDDVSCSGKETRFLQCSRRQWGRHDCSHRED 480
Query: 752 AGVYCAGNASCGEVYTEERNQLS-SFGYNTTFGANPWQVRIIPRVANEWCSGTIINKNW 925
+ C E ++LS F G N + R+ + +W GTI + W
Sbjct: 481 VSIACYPGG--------EGHRLSLGFPVRLVDGENKKEGRVEVFINGQW--GTICDDGW 529
Score = 39.7 bits (91), Expect = 0.025
Identities = 26/104 (25%), Positives = 40/104 (38%), Gaps = 4/104 (3%)
Frame = +2
Query: 194 KPGHFQGYMLYNDSMICMDSFAQNNAEVYCRYLGYPGARKFXXXXXXXXXXXXLF--NLQ 367
K G + ++ IC D + +A V CR LGY G + + N++
Sbjct: 509 KEGRVEVFINGQWGTICDDGWTDKDAAVICRQLGYKGPARARTMAYFGEGKGPIHVDNVK 568
Query: 368 CNGNENSPAEC--RVYTNSYCTSGRALGTVCQTVALAGTNRSNE 493
C GNE S A+C + C G +C + SN+
Sbjct: 569 CTGNERSLADCIKQDIGRHNCRHSEDAGVICDYFGKKASGNSNK 612
>sp|Q5G268|NETR_HYLLE Neurotrypsin precursor Length = 875 Score = 141 bits (356), Expect = 5e-33 Identities = 126/477 (26%), Positives = 192/477 (40%), Gaps = 45/477 (9%) Frame = +2 Query: 200 GHFQGYMLYNDSMICMDSFAQNNAEVYCRYLGYP-GARKFXXXXXXXXXXXXLFNLQCNG 376 G + Y +C D + + N V CR LG+ G + L ++ C+G Sbjct: 398 GRLEVYYRGQWGTVCDDGWTELNTYVVCRQLGFKYGKQASANHFEESTGPIWLDDVSCSG 457 Query: 377 NENSPAEC--RVYTNSYCTSGRALGTVCQT------------VALAGTNRSNEGRIEVYN 514 E +C R + C+ + C V L EGR+EV+ Sbjct: 458 KETRFLQCSRRQWGRHDCSHREDVSIACYPGGEGHRLSLGFPVRLMDGENKKEGRVEVFI 517 Query: 515 AGQKGLMCDDTFNADIANIVCRQLGFSFALNQTTAAGYVYGFGNKNFLLDYLYCNGNEDS 694 GQ G +CDD + A ++CRQLG+ T A +G G +D + C GNE S Sbjct: 518 NGQWGTICDDGWTDKDAAVICRQLGYKGPARARTMA--YFGEGKGPIHVDNVKCTGNERS 575 Query: 695 IDDCPWQYWRLRNCYGNDVAGVYC-------AGNAS-------CGEVYTEERNQLSSFGY 832 + DC Q NC ++ AGV C +GN++ CG R + G Sbjct: 576 LADCIKQDIGRHNCRHSEDAGVICDYFGKKASGNSNKESLSSVCGLRLLHRRQKRIIGGK 635 Query: 833 NTTFGANPWQVRIIPRVANE----WCSGTIINKNWVLTSANCFTMSYDNFXXXXXXXXXX 1000 N+ G PWQV + + ++ C T+++ WVLT+A+CF ++ Sbjct: 636 NSLRGGWPWQVSLRLKSSHGDGRLLCGATLLSSCWVLTAAHCFKRYGNSTRNYAVRVGDY 695 Query: 1001 XXXXNWAYTNNI--NSVFIHPQYSAVTGDYNLALVKTSP-----VAFDINVQPACLPSKN 1159 + I + IH +Y + DY++ALV+ F +V PACLP Sbjct: 696 HTLVPEEFEEEIGVQQIVIHREYRPDSSDYDIALVRLQGPEEQCARFSSHVLPACLPLWR 755 Query: 1160 LVP--NSLDCITAGWNHKLETNSRPLLGKSVNLYNTTNCKSLTQFDDDKLARYFCSDNNM 1333 P + +C GW SR L ++ L C+ ++ R C+ N Sbjct: 756 ERPQKTASNCYITGWGDTGRAYSRTLQQAAIPLLPKRFCEE--RYKGRFTGRMLCAGNLH 813 Query: 1334 GSSAI--FNGESGGPLICSIDG-SKVIAGVVSKSINSARATSIRLFTRVDYMLPWIK 1495 + G+SGGPL+C G S V+ GV S S ++T+V +PWIK Sbjct: 814 EHKRVDSCQGDSGGPLMCERPGESWVVYGVTSWGHGCGVKDSPGVYTKVSAFVPWIK 870
Score = 91.3 bits (225), Expect = 7e-18
Identities = 66/228 (28%), Positives = 100/228 (43%), Gaps = 12/228 (5%)
Frame = +2
Query: 119 INTNYFYCYSGSLQLKCQDKLQFTLKPGHFQGYMLYNDSMICMDSFAQNNAEVYCRYL-- 292
++ Y C GS++L+ K +F G + Y +C + ++A V C L
Sbjct: 158 VDWGYCDCRHGSIRLR-GGKNEFE---GTVEVYASGVWGTVCSSHWDDSDASVICHQLQL 213
Query: 293 GYPGARKFXXXXXXXXXXXXLFNLQCNGNENSPAECR--VYTNSYCTSGRALGTVCQ--- 457
G G K N++C G+E + C ++ C A C
Sbjct: 214 GGKGIAKQTPFSGLGLIPIYWSNVRCRGDEENILLCEKDIWQGGVCPQKMAAAVTCSFSH 273
Query: 458 -----TVALAGTNRSNEGRIEVYNAGQKGLMCDDTFNADIANIVCRQLGFSFALNQTTAA 622
+ L G + +EGR+E+Y+AGQ G +CDD ++ A ++CRQLG S A
Sbjct: 274 GPTFPIIRLVGGSSVHEGRVELYHAGQWGTVCDDQWDDADAEVICRQLGLSGIAKAWHQA 333
Query: 623 GYVYGFGNKNFLLDYLYCNGNEDSIDDCPWQYWRLRNCYGNDVAGVYC 766
+G G+ +LD + C GNE SI+ CP W NC + AGV C
Sbjct: 334 --YFGEGSGPVMLDEVRCTGNELSIEQCPKSSWGEHNCGHKEDAGVSC 379
Score = 80.5 bits (197), Expect = 1e-14
Identities = 68/239 (28%), Positives = 97/239 (40%), Gaps = 10/239 (4%)
Frame = +2
Query: 239 ICMDSFAQNNAEVYCRYLGYPGARK--FXXXXXXXXXXXXLFNLQCNGNENSPAEC--RV 406
+C D + +AEV CR LG G K L ++C GNE S +C
Sbjct: 304 VCDDQWDDADAEVICRQLGLSGIAKAWHQAYFGEGSGPVMLDEVRCTGNELSIEQCPKSS 363
Query: 407 YTNSYCTSGRALGTVCQ-----TVALAGTNRSNEGRIEVYNAGQKGLMCDDTFNADIANI 571
+ C G C + LAG S+EGR+EVY GQ G +CDD + +
Sbjct: 364 WGEHNCGHKEDAGVSCTPLTDGVIRLAGGKGSHEGRLEVYYRGQWGTVCDDGWTELNTYV 423
Query: 572 VCRQLGFSFALNQTTAAGYVYGFGNKNFLLDYLYCNGNEDSIDDCPWQYWRLRNCYGNDV 751
VCRQLGF + Q +A + G LD + C+G E C + W +C +
Sbjct: 424 VCRQLGFKYG-KQASANHFEESTG--PIWLDDVSCSGKETRFLQCSRRQWGRHDCSHRED 480
Query: 752 AGVYCAGNASCGEVYTEERNQLS-SFGYNTTFGANPWQVRIIPRVANEWCSGTIINKNW 925
+ C E ++LS F G N + R+ + +W GTI + W
Sbjct: 481 VSIACYPGG--------EGHRLSLGFPVRLMDGENKKEGRVEVFINGQW--GTICDDGW 529
Score = 39.7 bits (91), Expect = 0.025
Identities = 26/104 (25%), Positives = 40/104 (38%), Gaps = 4/104 (3%)
Frame = +2
Query: 194 KPGHFQGYMLYNDSMICMDSFAQNNAEVYCRYLGYPGARKFXXXXXXXXXXXXLF--NLQ 367
K G + ++ IC D + +A V CR LGY G + + N++
Sbjct: 509 KEGRVEVFINGQWGTICDDGWTDKDAAVICRQLGYKGPARARTMAYFGEGKGPIHVDNVK 568
Query: 368 CNGNENSPAEC--RVYTNSYCTSGRALGTVCQTVALAGTNRSNE 493
C GNE S A+C + C G +C + SN+
Sbjct: 569 CTGNERSLADCIKQDIGRHNCRHSEDAGVICDYFGKKASGNSNK 612
>sp|Q5G270|NETR_GORGO Neurotrypsin precursor Length = 876 Score = 141 bits (355), Expect = 6e-33 Identities = 126/477 (26%), Positives = 190/477 (39%), Gaps = 45/477 (9%) Frame = +2 Query: 200 GHFQGYMLYNDSMICMDSFAQNNAEVYCRYLGYP-GARKFXXXXXXXXXXXXLFNLQCNG 376 G + Y +C D + + N V CR LG+ G + L ++ C+G Sbjct: 399 GRLEVYYRGQWGTVCDDGWTELNTYVVCRQLGFKYGKQASANHFEESTGPIWLDDVSCSG 458 Query: 377 NENSPAEC--RVYTNSYCTSGRALGTVCQT------------VALAGTNRSNEGRIEVYN 514 E +C R + C+ + C V L EGR+EV+ Sbjct: 459 KETRFLQCSRRQWGRHDCSHREDVSIACYPGGEGHRLSLGFPVRLMDGENKKEGRVEVFI 518 Query: 515 AGQKGLMCDDTFNADIANIVCRQLGFSFALNQTTAAGYVYGFGNKNFLLDYLYCNGNEDS 694 GQ G +CDD + A ++CRQLG+ T A +G G +D + C GNE S Sbjct: 519 NGQWGTICDDGWTDKDAAVICRQLGYKGPARARTMA--YFGEGKGPIHVDNVKCTGNERS 576 Query: 695 IDDCPWQYWRLRNCYGNDVAGVYC-------AGNAS-------CGEVYTEERNQLSSFGY 832 + DC Q NC ++ AGV C +GN++ CG R + G Sbjct: 577 LADCIKQDIGRHNCRHSEDAGVICDYFGKKASGNSNKESLSSVCGLRLLHRRQKRIIGGK 636 Query: 833 NTTFGANPWQVRIIPRV----ANEWCSGTIINKNWVLTSANCFTMSYDNFXXXXXXXXXX 1000 N+ G PWQV + + C T+++ WVLT+A+CF ++ Sbjct: 637 NSLRGGWPWQVSLRLKSYHGDGRLLCGATLLSSCWVLTAAHCFKRYGNSTRNYAVRVGDY 696 Query: 1001 XXXXNWAYTNNI--NSVFIHPQYSAVTGDYNLALVKTSP-----VAFDINVQPACLPSKN 1159 + I + IH +Y + DY++ALV+ F +V PACLP Sbjct: 697 HTLVPEEFEEEIGVQQIVIHREYRPDSSDYDIALVRLQGPEEQCARFSSHVLPACLPLWR 756 Query: 1160 LVP--NSLDCITAGWNHKLETNSRPLLGKSVNLYNTTNCKSLTQFDDDKLARYFCSDNNM 1333 P + +C GW SR L ++ L C+ ++ R C+ N Sbjct: 757 ERPQKTASNCYITGWGDTGRAYSRTLQQAAIPLLPKRFCEE--RYKGRFTGRMLCAGNLH 814 Query: 1334 GSSAI--FNGESGGPLICSIDG-SKVIAGVVSKSINSARATSIRLFTRVDYMLPWIK 1495 + G+SGGPL+C G S V+ GV S S ++T+V +PWIK Sbjct: 815 EHKRVDSCQGDSGGPLMCERPGESWVVYGVTSWGYGCGVKDSPGVYTKVSAFVPWIK 871
Score = 92.4 bits (228), Expect = 3e-18
Identities = 67/228 (29%), Positives = 101/228 (44%), Gaps = 12/228 (5%)
Frame = +2
Query: 119 INTNYFYCYSGSLQLKCQDKLQFTLKPGHFQGYMLYNDSMICMDSFAQNNAEVYCRYL-- 292
++ Y C GS++L+ K +F G + Y +C + ++A V C L
Sbjct: 159 VDWGYCDCRHGSVRLR-GGKNEFE---GTVEVYASGVWGTVCSSHWDDSDASVICHQLQL 214
Query: 293 GYPGARKFXXXXXXXXXXXXLFNLQCNGNENSPAECR--VYTNSYCTSGRALGTVCQ--- 457
G G K N++C G+E + C ++ C A C
Sbjct: 215 GGKGIAKQTPFSGLGLIPVYWSNVRCRGDEENILLCEKDIWQGGVCPQKMAAAVTCSFSH 274
Query: 458 -----TVALAGTNRSNEGRIEVYNAGQKGLMCDDTFNADIANIVCRQLGFSFALNQTTAA 622
+ LAG + +EGR+E+Y+AGQ G +CDD ++ A ++CRQLG S A
Sbjct: 275 GPTFPIIRLAGGSSVHEGRVELYHAGQWGTVCDDQWDDADAEVICRQLGLSGIAKAWHQA 334
Query: 623 GYVYGFGNKNFLLDYLYCNGNEDSIDDCPWQYWRLRNCYGNDVAGVYC 766
+G G+ +LD + C GNE SI+ CP W NC + AGV C
Sbjct: 335 --YFGEGSGPVMLDEVRCTGNELSIEQCPKSSWGEHNCGHKEDAGVSC 380
Score = 80.5 bits (197), Expect = 1e-14
Identities = 68/239 (28%), Positives = 97/239 (40%), Gaps = 10/239 (4%)
Frame = +2
Query: 239 ICMDSFAQNNAEVYCRYLGYPGARK--FXXXXXXXXXXXXLFNLQCNGNENSPAEC--RV 406
+C D + +AEV CR LG G K L ++C GNE S +C
Sbjct: 305 VCDDQWDDADAEVICRQLGLSGIAKAWHQAYFGEGSGPVMLDEVRCTGNELSIEQCPKSS 364
Query: 407 YTNSYCTSGRALGTVCQ-----TVALAGTNRSNEGRIEVYNAGQKGLMCDDTFNADIANI 571
+ C G C + LAG S+EGR+EVY GQ G +CDD + +
Sbjct: 365 WGEHNCGHKEDAGVSCTPLTDGVIRLAGGKGSHEGRLEVYYRGQWGTVCDDGWTELNTYV 424
Query: 572 VCRQLGFSFALNQTTAAGYVYGFGNKNFLLDYLYCNGNEDSIDDCPWQYWRLRNCYGNDV 751
VCRQLGF + Q +A + G LD + C+G E C + W +C +
Sbjct: 425 VCRQLGFKYG-KQASANHFEESTG--PIWLDDVSCSGKETRFLQCSRRQWGRHDCSHRED 481
Query: 752 AGVYCAGNASCGEVYTEERNQLS-SFGYNTTFGANPWQVRIIPRVANEWCSGTIINKNW 925
+ C E ++LS F G N + R+ + +W GTI + W
Sbjct: 482 VSIACYPGG--------EGHRLSLGFPVRLMDGENKKEGRVEVFINGQW--GTICDDGW 530
Score = 39.7 bits (91), Expect = 0.025
Identities = 26/104 (25%), Positives = 40/104 (38%), Gaps = 4/104 (3%)
Frame = +2
Query: 194 KPGHFQGYMLYNDSMICMDSFAQNNAEVYCRYLGYPGARKFXXXXXXXXXXXXLF--NLQ 367
K G + ++ IC D + +A V CR LGY G + + N++
Sbjct: 510 KEGRVEVFINGQWGTICDDGWTDKDAAVICRQLGYKGPARARTMAYFGEGKGPIHVDNVK 569
Query: 368 CNGNENSPAEC--RVYTNSYCTSGRALGTVCQTVALAGTNRSNE 493
C GNE S A+C + C G +C + SN+
Sbjct: 570 CTGNERSLADCIKQDIGRHNCRHSEDAGVICDYFGKKASGNSNK 613
>sp|Q5G267|NETR_MACMU Neurotrypsin precursor Length = 875 Score = 140 bits (354), Expect = 8e-33 Identities = 125/477 (26%), Positives = 191/477 (40%), Gaps = 45/477 (9%) Frame = +2 Query: 200 GHFQGYMLYNDSMICMDSFAQNNAEVYCRYLGYP-GARKFXXXXXXXXXXXXLFNLQCNG 376 G + Y +C D + + N V CR LG+ G + L ++ C+G Sbjct: 398 GRLEVYYRGQWGTVCDDGWTELNTYVVCRQLGFKYGKQASANHFEESTGPIWLDDVSCSG 457 Query: 377 NENSPAEC--RVYTNSYCTSGRALGTVCQT------------VALAGTNRSNEGRIEVYN 514 E +C R + C+ + C V L EGR+EV+ Sbjct: 458 KETRFLQCSRRQWGQHDCSHHEDVSIACYPGSEGHRLSLGFPVRLMDGENKKEGRVEVFI 517 Query: 515 AGQKGLMCDDTFNADIANIVCRQLGFSFALNQTTAAGYVYGFGNKNFLLDYLYCNGNEDS 694 GQ G +CDD + A ++CRQLG+ T A +G G +D + C GNE S Sbjct: 518 NGQWGTICDDGWTDKDAAVICRQLGYKGPARARTMA--YFGEGKGPIHVDNVKCTGNERS 575 Query: 695 IDDCPWQYWRLRNCYGNDVAGVYC-------AGNAS-------CGEVYTEERNQLSSFGY 832 + DC Q NC ++ AGV C +GN++ CG R + G Sbjct: 576 LADCIKQDIGRHNCRHSEDAGVICDYFGKKASGNSNKESLSSVCGLRLLHRRQKRIIGGK 635 Query: 833 NTTFGANPWQVRIIPRVANE----WCSGTIINKNWVLTSANCFTMSYDNFXXXXXXXXXX 1000 N+ G PWQV + + ++ C T+++ WVLT+A+CF ++ Sbjct: 636 NSLRGGWPWQVSLRLKSSHGDGRLLCGATLLSSCWVLTAAHCFKRYGNSTRNYAVRVGDY 695 Query: 1001 XXXXNWAYTNNI--NSVFIHPQYSAVTGDYNLALVKTSP-----VAFDINVQPACLPSKN 1159 + I + IH +Y + DY++ALV+ F +V PACLP Sbjct: 696 HTLVPEEFEEEIGVQQIVIHREYRPDSSDYDIALVRLQGPEEQCARFSSHVLPACLPFWR 755 Query: 1160 LVP--NSLDCITAGWNHKLETNSRPLLGKSVNLYNTTNCKSLTQFDDDKLARYFCSDNNM 1333 P + +C GW SR L ++ L C+ ++ R C+ N Sbjct: 756 ERPQKTASNCYITGWGDTGRAYSRTLQQAAIPLLPKRFCEE--RYKGRFTGRMLCAGNLH 813 Query: 1334 GSSAI--FNGESGGPLICSIDG-SKVIAGVVSKSINSARATSIRLFTRVDYMLPWIK 1495 + G+SGGPL+C G S + GV S S ++T+V +PWIK Sbjct: 814 EHKRVDSCQGDSGGPLMCERPGESWAVYGVTSWGYGCGVKDSPGVYTKVSAFVPWIK 870
Score = 91.3 bits (225), Expect = 7e-18
Identities = 66/228 (28%), Positives = 100/228 (43%), Gaps = 12/228 (5%)
Frame = +2
Query: 119 INTNYFYCYSGSLQLKCQDKLQFTLKPGHFQGYMLYNDSMICMDSFAQNNAEVYCRYL-- 292
++ Y C GS++L+ K +F G + Y +C + ++A V C L
Sbjct: 158 VDWGYCDCRHGSVRLR-GGKNEFE---GTVEVYASGAWGTVCSSHWDDSDASVICHQLQL 213
Query: 293 GYPGARKFXXXXXXXXXXXXLFNLQCNGNENSPAECR--VYTNSYCTSGRALGTVCQ--- 457
G G K N++C G+E + C ++ C A C
Sbjct: 214 GGKGIAKQTPFSGLGLIPIYWSNVRCRGDEENILLCEKDIWQGGVCPQKMAAAVTCSFSR 273
Query: 458 -----TVALAGTNRSNEGRIEVYNAGQKGLMCDDTFNADIANIVCRQLGFSFALNQTTAA 622
+ L G + +EGR+E+Y+AGQ G +CDD ++ A ++CRQLG S A
Sbjct: 274 GPAFPIIRLVGGSSVHEGRVELYHAGQWGTICDDQWDDADAEVICRQLGLSGIAKAWHQA 333
Query: 623 GYVYGFGNKNFLLDYLYCNGNEDSIDDCPWQYWRLRNCYGNDVAGVYC 766
+G G+ +LD + C GNE SI+ CP W NC + AGV C
Sbjct: 334 --YFGEGSGPVMLDEVRCTGNELSIEQCPKSSWGEHNCGHKEDAGVSC 379
Score = 81.6 bits (200), Expect = 6e-15
Identities = 69/239 (28%), Positives = 99/239 (41%), Gaps = 10/239 (4%)
Frame = +2
Query: 239 ICMDSFAQNNAEVYCRYLGYPGARK--FXXXXXXXXXXXXLFNLQCNGNENSPAEC--RV 406
IC D + +AEV CR LG G K L ++C GNE S +C
Sbjct: 304 ICDDQWDDADAEVICRQLGLSGIAKAWHQAYFGEGSGPVMLDEVRCTGNELSIEQCPKSS 363
Query: 407 YTNSYCTSGRALGTVCQ-----TVALAGTNRSNEGRIEVYNAGQKGLMCDDTFNADIANI 571
+ C G C + LAG S+EGR+EVY GQ G +CDD + +
Sbjct: 364 WGEHNCGHKEDAGVSCTPLTDGVIRLAGGKGSHEGRLEVYYRGQWGTVCDDGWTELNTYV 423
Query: 572 VCRQLGFSFALNQTTAAGYVYGFGNKNFLLDYLYCNGNEDSIDDCPWQYWRLRNCYGNDV 751
VCRQLGF + Q +A + G LD + C+G E C + W +C ++
Sbjct: 424 VCRQLGFKYG-KQASANHFEESTG--PIWLDDVSCSGKETRFLQCSRRQWGQHDCSHHED 480
Query: 752 AGVYCAGNASCGEVYTEERNQLS-SFGYNTTFGANPWQVRIIPRVANEWCSGTIINKNW 925
+ C + E ++LS F G N + R+ + +W GTI + W
Sbjct: 481 VSIACYPGS--------EGHRLSLGFPVRLMDGENKKEGRVEVFINGQW--GTICDDGW 529
Score = 39.7 bits (91), Expect = 0.025
Identities = 26/104 (25%), Positives = 40/104 (38%), Gaps = 4/104 (3%)
Frame = +2
Query: 194 KPGHFQGYMLYNDSMICMDSFAQNNAEVYCRYLGYPGARKFXXXXXXXXXXXXLF--NLQ 367
K G + ++ IC D + +A V CR LGY G + + N++
Sbjct: 509 KEGRVEVFINGQWGTICDDGWTDKDAAVICRQLGYKGPARARTMAYFGEGKGPIHVDNVK 568
Query: 368 CNGNENSPAEC--RVYTNSYCTSGRALGTVCQTVALAGTNRSNE 493
C GNE S A+C + C G +C + SN+
Sbjct: 569 CTGNERSLADCIKQDIGRHNCRHSEDAGVICDYFGKKASGNSNK 612
>sp|Q5G266|NETR_TRAPH Neurotrypsin precursor Length = 875 Score = 140 bits (353), Expect = 1e-32 Identities = 125/477 (26%), Positives = 191/477 (40%), Gaps = 45/477 (9%) Frame = +2 Query: 200 GHFQGYMLYNDSMICMDSFAQNNAEVYCRYLGYP-GARKFXXXXXXXXXXXXLFNLQCNG 376 G + Y +C D + + N V CR LG+ G + L ++ C+G Sbjct: 398 GRLEVYYRGQWGTVCDDGWTELNTYVACRQLGFKYGKQASANHFEESTGPIWLDDVSCSG 457 Query: 377 NENSPAEC--RVYTNSYCTSGRALGTVCQT------------VALAGTNRSNEGRIEVYN 514 E +C R + C+ + C V L EGR+EV+ Sbjct: 458 KETRFLQCSRRQWGRHDCSHREDVSIACYPGSEGHRLSLGFPVRLMDGENKKEGRVEVFI 517 Query: 515 AGQKGLMCDDTFNADIANIVCRQLGFSFALNQTTAAGYVYGFGNKNFLLDYLYCNGNEDS 694 GQ G +CDD + A ++CRQLG+ T A +G G +D + C GNE S Sbjct: 518 NGQWGTICDDGWTDKDAAVICRQLGYKGPARARTMA--YFGEGKGPIHVDNVKCTGNERS 575 Query: 695 IDDCPWQYWRLRNCYGNDVAGVYC-------AGNAS-------CGEVYTEERNQLSSFGY 832 + DC Q NC ++ AGV C +GN++ CG R + G Sbjct: 576 LADCIKQDIGRHNCRHSEDAGVICDYFGKKASGNSNKESLSSVCGLRLLHRRQKRIIGGK 635 Query: 833 NTTFGANPWQVRIIPRVANE----WCSGTIINKNWVLTSANCFTMSYDNFXXXXXXXXXX 1000 N+ G PWQV + + ++ C T+++ WVLT+A+CF ++ Sbjct: 636 NSLRGGWPWQVSLRLKSSHGDGRLLCGATLLSSCWVLTAAHCFKRYGNSTRNYAVRVGDY 695 Query: 1001 XXXXNWAYTNNI--NSVFIHPQYSAVTGDYNLALVKTSP-----VAFDINVQPACLPSKN 1159 + I + IH +Y + DY++ALV+ F +V PACLP Sbjct: 696 HTLVPEEFEEEIGVQQIVIHREYRPDSSDYDIALVRLQGPEEQCARFSSHVLPACLPFWR 755 Query: 1160 LVP--NSLDCITAGWNHKLETNSRPLLGKSVNLYNTTNCKSLTQFDDDKLARYFCSDNNM 1333 P + +C GW SR L ++ L C+ ++ R C+ N Sbjct: 756 ERPQKTASNCYITGWGDTGRAYSRTLQQAAIPLLPKRFCEE--RYKGRFTGRMLCAGNLH 813 Query: 1334 GSSAI--FNGESGGPLICSIDG-SKVIAGVVSKSINSARATSIRLFTRVDYMLPWIK 1495 + G+SGGPL+C G S + GV S S ++T+V +PWIK Sbjct: 814 EHKRVDSCQGDSGGPLMCERPGESWAVYGVTSWGYGCGVKDSPGVYTKVSAFVPWIK 870
Score = 92.4 bits (228), Expect = 3e-18
Identities = 67/228 (29%), Positives = 100/228 (43%), Gaps = 12/228 (5%)
Frame = +2
Query: 119 INTNYFYCYSGSLQLKCQDKLQFTLKPGHFQGYMLYNDSMICMDSFAQNNAEVYCRYL-- 292
++ Y C GS++L+ K +F G + Y +C + ++A V C L
Sbjct: 158 VDWGYCDCRHGSVRLR-GGKNEFE---GTVEVYASGVWGTVCSSHWDDSDASVICHQLQL 213
Query: 293 GYPGARKFXXXXXXXXXXXXLFNLQCNGNENSPAECR--VYTNSYCTSGRALGTVCQ--- 457
G G K N++C G+E + C ++ C A C
Sbjct: 214 GGKGIAKQTPFSGLGLIPIYWSNVRCRGDEENILLCEKDIWQXGVCPQKMAAAVTCSFSH 273
Query: 458 -----TVALAGTNRSNEGRIEVYNAGQKGLMCDDTFNADIANIVCRQLGFSFALNQTTAA 622
+ L G N +EGR+E+Y+AGQ G +CDD ++ A ++CRQLG S A
Sbjct: 274 GPAFPIIRLVGGNSVHEGRVELYHAGQWGTVCDDQWDDADAEVICRQLGLSGIAKAWHQA 333
Query: 623 GYVYGFGNKNFLLDYLYCNGNEDSIDDCPWQYWRLRNCYGNDVAGVYC 766
+G G+ +LD + C GNE SI+ CP W NC + AGV C
Sbjct: 334 --YFGEGSGPVMLDEVRCTGNELSIEQCPKSSWGEHNCGHKEDAGVSC 379
Score = 79.3 bits (194), Expect = 3e-14
Identities = 67/239 (28%), Positives = 97/239 (40%), Gaps = 10/239 (4%)
Frame = +2
Query: 239 ICMDSFAQNNAEVYCRYLGYPGARK--FXXXXXXXXXXXXLFNLQCNGNENSPAEC--RV 406
+C D + +AEV CR LG G K L ++C GNE S +C
Sbjct: 304 VCDDQWDDADAEVICRQLGLSGIAKAWHQAYFGEGSGPVMLDEVRCTGNELSIEQCPKSS 363
Query: 407 YTNSYCTSGRALGTVCQ-----TVALAGTNRSNEGRIEVYNAGQKGLMCDDTFNADIANI 571
+ C G C + LAG S+EGR+EVY GQ G +CDD + +
Sbjct: 364 WGEHNCGHKEDAGVSCTPLTDGVIRLAGGKGSHEGRLEVYYRGQWGTVCDDGWTELNTYV 423
Query: 572 VCRQLGFSFALNQTTAAGYVYGFGNKNFLLDYLYCNGNEDSIDDCPWQYWRLRNCYGNDV 751
CRQLGF + Q +A + G LD + C+G E C + W +C +
Sbjct: 424 ACRQLGFKYG-KQASANHFEESTG--PIWLDDVSCSGKETRFLQCSRRQWGRHDCSHRED 480
Query: 752 AGVYCAGNASCGEVYTEERNQLS-SFGYNTTFGANPWQVRIIPRVANEWCSGTIINKNW 925
+ C + E ++LS F G N + R+ + +W GTI + W
Sbjct: 481 VSIACYPGS--------EGHRLSLGFPVRLMDGENKKEGRVEVFINGQW--GTICDDGW 529
Score = 39.7 bits (91), Expect = 0.025
Identities = 26/104 (25%), Positives = 40/104 (38%), Gaps = 4/104 (3%)
Frame = +2
Query: 194 KPGHFQGYMLYNDSMICMDSFAQNNAEVYCRYLGYPGARKFXXXXXXXXXXXXLF--NLQ 367
K G + ++ IC D + +A V CR LGY G + + N++
Sbjct: 509 KEGRVEVFINGQWGTICDDGWTDKDAAVICRQLGYKGPARARTMAYFGEGKGPIHVDNVK 568
Query: 368 CNGNENSPAEC--RVYTNSYCTSGRALGTVCQTVALAGTNRSNE 493
C GNE S A+C + C G +C + SN+
Sbjct: 569 CTGNERSLADCIKQDIGRHNCRHSEDAGVICDYFGKKASGNSNK 612
>sp|Q9BYE2|TMPSD_HUMAN Transmembrane protease, serine 13 (Mosaic serine protease) (Membrane-type mosaic serine protease) Length = 581 Score = 99.4 bits (246), Expect = 3e-20 Identities = 82/325 (25%), Positives = 131/325 (40%), Gaps = 5/325 (1%) Frame = +2 Query: 533 MCDDTFNADIANIVCRQLGFSFALNQTTAAGYVYGFGNKNFLLDYLYCNGNEDSIDDCPW 712 +C +N + C+QLGF A T A F N +L Y +CP Sbjct: 244 ICSSNWNDSYSEKTCQQLGFESAHRTTEVAHR--DFANSFSILRYNSTIQESLHRSECPS 301 Query: 713 QYWRLRNCYGNDVAGVYCAGNASCGEVYTEERNQLSSFGYNTTFGANPWQVRIIPRVANE 892 Q + C +C A G + S + PWQV + + Sbjct: 302 QRYISLQCS-------HCGLRAMTGRIVGGALASDSKW---------PWQVSLHFGTTHI 345 Query: 893 WCSGTIINKNWVLTSANCFTMSYDNFXXXXXXXXXXXXXXNWAYTNNINSVFIHPQYSAV 1072 C GT+I+ WVLT+A+CF ++ + +I + I+ Y+ Sbjct: 346 -CGGTLIDAQWVLTAAHCFFVTREKVLEGWKVYAGTSNLHQLPEAASIAEIIINSNYTDE 404 Query: 1073 TGDYNLALVKTS-PVAFDINVQPACLPSKNLVPNSLD-CITAGWNHKLETNSR--PLLGK 1240 DY++AL++ S P+ ++ PACLP + + C G+ ET+ + P L + Sbjct: 405 EDDYDIALMRLSKPLTLSAHIHPACLPMHGQTFSLNETCWITGFGKTRETDDKTSPFLRE 464 Query: 1241 -SVNLYNTTNCKSLTQFDDDKLARYFCSDNNMGSSAIFNGESGGPLICSIDGSKVIAGVV 1417 VNL + C +D R C+ + G G+SGGPL+C + +AGV Sbjct: 465 VQVNLIDFKKCNDYLVYDSYLTPRMMCAGDLHGGRDSCQGDSGGPLVCEQNNRWYLAGVT 524 Query: 1418 SKSINSARATSIRLFTRVDYMLPWI 1492 S + ++T+V +LPWI Sbjct: 525 SWGTGCGQRNKPGVYTKVTEVLPWI 549
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 180,935,846
Number of Sequences: 369166
Number of extensions: 3942820
Number of successful extensions: 10600
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 9026
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 9898
length of database: 68,354,980
effective HSP length: 115
effective length of database: 47,110,455
effective search space used: 19645059735
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail