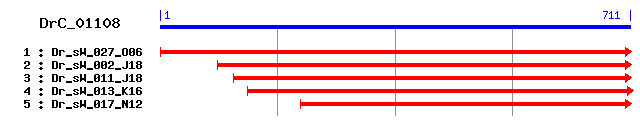
DrC_01108
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_01108 (711 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P09671|SODM_MOUSE Superoxide dismutase [Mn], mitochondri... 298 1e-80 sp|P07895|SODM_RAT Superoxide dismutase [Mn], mitochondrial... 296 4e-80 sp|P41976|SODM_BOVIN Superoxide dismutase [Mn], mitochondri... 295 6e-80 sp|Q8HXP4|SODM_MACFU Superoxide dismutase [Mn], mitochondri... 295 6e-80 sp|Q8HXP6|SODM_PONPY Superoxide dismutase [Mn], mitochondri... 294 2e-79 sp|P04179|SODM_HUMAN Superoxide dismutase [Mn], mitochondri... 294 2e-79 sp|Q8HXP7|SODM_PANTR Superoxide dismutase [Mn], mitochondrial 294 2e-79 sp|Q8HXP1|SODM_CEBAP Superoxide dismutase [Mn], mitochondri... 294 2e-79 sp|Q5FB30|SODM_MACNE Superoxide dismutase [Mn], mitochondri... 293 2e-79 sp|Q8HXP5|SODM_HYLLA Superoxide dismutase [Mn], mitochondrial 292 5e-79
>sp|P09671|SODM_MOUSE Superoxide dismutase [Mn], mitochondrial precursor Length = 222 Score = 298 bits (762), Expect = 1e-80 Identities = 130/194 (67%), Positives = 158/194 (81%) Frame = +2 Query: 53 KHTLPDLPYDYNALEPSISADIMEVHHKKHHAAYVNNLNSALEQLDEAVHKNDVNKIISM 232 KH+LPDLPYDY ALEP I+A IM++HH KHHAAYVNNLN+ E+ EA+ K DV +++ Sbjct: 25 KHSLPDLPYDYGALEPHINAQIMQLHHSKHHAAYVNNLNATEEKYHEALAKGDVTTQVAL 84 Query: 233 QNAIKFNGGGHLNHSIFWKNLCPKGGGEPDGSLLAALQREFGSFENFKTELSAAAVGIQG 412 Q A+KFNGGGH+NH+IFW NL PKGGGEP G LL A++R+FGSFE FK +L+A +VG+QG Sbjct: 85 QPALKFNGGGHINHTIFWTNLSPKGGGEPKGELLEAIKRDFGSFEKFKEKLTAVSVGVQG 144 Query: 413 SGWAWLGLNKKNGRLHITTCGNQDPLEATTGLKPLLGFDVWEHAYYLQYKNVRPDYVKAL 592 SGW WLG NK+ GRL I C NQDPL+ TTGL PLLG DVWEHAYYLQYKNVRPDY+KA+ Sbjct: 145 SGWGWLGFNKEQGRLQIAACSNQDPLQGTTGLIPLLGIDVWEHAYYLQYKNVRPDYLKAI 204 Query: 593 WRIINWGDVAHRFS 634 W +INW +V R++ Sbjct: 205 WNVINWENVTERYT 218
>sp|P07895|SODM_RAT Superoxide dismutase [Mn], mitochondrial precursor Length = 222 Score = 296 bits (758), Expect = 4e-80 Identities = 130/193 (67%), Positives = 156/193 (80%) Frame = +2 Query: 53 KHTLPDLPYDYNALEPSISADIMEVHHKKHHAAYVNNLNSALEQLDEAVHKNDVNKIISM 232 KH+LPDLPYDY ALEP I+A IM++HH KHHA YVNNLN E+ EA+ K DV +++ Sbjct: 25 KHSLPDLPYDYGALEPHINAQIMQLHHSKHHATYVNNLNVTEEKYHEALAKGDVTTQVAL 84 Query: 233 QNAIKFNGGGHLNHSIFWKNLCPKGGGEPDGSLLAALQREFGSFENFKTELSAAAVGIQG 412 Q A+KFNGGGH+NHSIFW NL PKGGGEP G LL A++R+FGSFE FK +L+A +VG+QG Sbjct: 85 QPALKFNGGGHINHSIFWTNLSPKGGGEPKGELLEAIKRDFGSFEKFKEKLTAVSVGVQG 144 Query: 413 SGWAWLGLNKKNGRLHITTCGNQDPLEATTGLKPLLGFDVWEHAYYLQYKNVRPDYVKAL 592 SGW WLG NK+ GRL I C NQDPL+ TTGL PLLG DVWEHAYYLQYKNVRPDY+KA+ Sbjct: 145 SGWGWLGFNKEQGRLQIAACSNQDPLQGTTGLIPLLGIDVWEHAYYLQYKNVRPDYLKAI 204 Query: 593 WRIINWGDVAHRF 631 W +INW +V+ R+ Sbjct: 205 WNVINWENVSQRY 217
>sp|P41976|SODM_BOVIN Superoxide dismutase [Mn], mitochondrial precursor Length = 222 Score = 295 bits (756), Expect = 6e-80 Identities = 131/194 (67%), Positives = 156/194 (80%) Frame = +2 Query: 53 KHTLPDLPYDYNALEPSISADIMEVHHKKHHAAYVNNLNSALEQLDEAVHKNDVNKIISM 232 KH+LPDLPYDY ALEP I+A IM++HH KHHAAYVNNLN A E+ EA+ K DV I++ Sbjct: 25 KHSLPDLPYDYGALEPHINAQIMQLHHSKHHAAYVNNLNVAEEKYREALEKGDVTAQIAL 84 Query: 233 QNAIKFNGGGHLNHSIFWKNLCPKGGGEPDGSLLAALQREFGSFENFKTELSAAAVGIQG 412 Q A+KFNGGGH+NHSIFW NL P GGGEP G LL A++R+FGSF FK +L+A +VG+QG Sbjct: 85 QPALKFNGGGHINHSIFWTNLSPNGGGEPQGELLEAIKRDFGSFAKFKEKLTAVSVGVQG 144 Query: 413 SGWAWLGLNKKNGRLHITTCGNQDPLEATTGLKPLLGFDVWEHAYYLQYKNVRPDYVKAL 592 SGW WLG NK+ GRL I C NQDPL+ TTGL PLLG DVWEHAYYLQYKNVRPDY+KA+ Sbjct: 145 SGWGWLGFNKEQGRLQIAACSNQDPLQGTTGLIPLLGIDVWEHAYYLQYKNVRPDYLKAI 204 Query: 593 WRIINWGDVAHRFS 634 W +INW +V R++ Sbjct: 205 WNVINWENVTARYT 218
>sp|Q8HXP4|SODM_MACFU Superoxide dismutase [Mn], mitochondrial sp|Q8HXP3|SODM_MACFA Superoxide dismutase [Mn], mitochondrial sp|Q8HXP2|SODM_MACMU Superoxide dismutase [Mn], mitochondrial Length = 198 Score = 295 bits (756), Expect = 6e-80 Identities = 131/193 (67%), Positives = 156/193 (80%) Frame = +2 Query: 53 KHTLPDLPYDYNALEPSISADIMEVHHKKHHAAYVNNLNSALEQLDEAVHKNDVNKIISM 232 KH+LPDLPYDY ALEP I+A IM++HH KHHAAYVNNLN E+ EA+ K DV I++ Sbjct: 1 KHSLPDLPYDYGALEPHINAQIMQLHHSKHHAAYVNNLNVTEEKYQEALAKGDVTAQIAL 60 Query: 233 QNAIKFNGGGHLNHSIFWKNLCPKGGGEPDGSLLAALQREFGSFENFKTELSAAAVGIQG 412 Q A+KFNGGGH+NHSIFW NL P GGGEP G LL A++R+FGSFE FK +L+AA+VG+QG Sbjct: 61 QPALKFNGGGHINHSIFWTNLSPNGGGEPKGELLEAIKRDFGSFEKFKEKLTAASVGVQG 120 Query: 413 SGWAWLGLNKKNGRLHITTCGNQDPLEATTGLKPLLGFDVWEHAYYLQYKNVRPDYVKAL 592 SGW WLG NK+ G+L I C NQDPL+ TTGL PLLG DVWEHAYYLQYKNVRPDY+KA+ Sbjct: 121 SGWGWLGFNKERGQLQIAACPNQDPLQGTTGLIPLLGIDVWEHAYYLQYKNVRPDYLKAI 180 Query: 593 WRIINWGDVAHRF 631 W +INW +V R+ Sbjct: 181 WNVINWENVTERY 193
>sp|Q8HXP6|SODM_PONPY Superoxide dismutase [Mn], mitochondrial precursor Length = 222 Score = 294 bits (752), Expect = 2e-79 Identities = 130/193 (67%), Positives = 155/193 (80%) Frame = +2 Query: 53 KHTLPDLPYDYNALEPSISADIMEVHHKKHHAAYVNNLNSALEQLDEAVHKNDVNKIISM 232 KH+LPDLPYDY ALEP I+A IM++HH KHHAAYVNNLN E+ EA+ K DV I++ Sbjct: 25 KHSLPDLPYDYGALEPHINAQIMQLHHSKHHAAYVNNLNVTEEKYQEALAKGDVTAQIAL 84 Query: 233 QNAIKFNGGGHLNHSIFWKNLCPKGGGEPDGSLLAALQREFGSFENFKTELSAAAVGIQG 412 Q A+KFNGGGH+NHSIFW NL P GGGEP G LL A++R+FGSF+ FK +L+AA+VG+QG Sbjct: 85 QPALKFNGGGHINHSIFWTNLSPNGGGEPKGELLEAIKRDFGSFDKFKEKLTAASVGVQG 144 Query: 413 SGWAWLGLNKKNGRLHITTCGNQDPLEATTGLKPLLGFDVWEHAYYLQYKNVRPDYVKAL 592 SGW WLG NK+ G L I C NQDPL+ TTGL PLLG DVWEHAYYLQYKNVRPDY+KA+ Sbjct: 145 SGWGWLGFNKERGHLQIAACPNQDPLQGTTGLIPLLGIDVWEHAYYLQYKNVRPDYLKAI 204 Query: 593 WRIINWGDVAHRF 631 W +INW +V R+ Sbjct: 205 WNVINWENVTERY 217
>sp|P04179|SODM_HUMAN Superoxide dismutase [Mn], mitochondrial precursor Length = 222 Score = 294 bits (752), Expect = 2e-79 Identities = 130/193 (67%), Positives = 155/193 (80%) Frame = +2 Query: 53 KHTLPDLPYDYNALEPSISADIMEVHHKKHHAAYVNNLNSALEQLDEAVHKNDVNKIISM 232 KH+LPDLPYDY ALEP I+A IM++HH KHHAAYVNNLN E+ EA+ K DV I++ Sbjct: 25 KHSLPDLPYDYGALEPHINAQIMQLHHSKHHAAYVNNLNVTEEKYQEALAKGDVTAQIAL 84 Query: 233 QNAIKFNGGGHLNHSIFWKNLCPKGGGEPDGSLLAALQREFGSFENFKTELSAAAVGIQG 412 Q A+KFNGGGH+NHSIFW NL P GGGEP G LL A++R+FGSF+ FK +L+AA+VG+QG Sbjct: 85 QPALKFNGGGHINHSIFWTNLSPNGGGEPKGELLEAIKRDFGSFDKFKEKLTAASVGVQG 144 Query: 413 SGWAWLGLNKKNGRLHITTCGNQDPLEATTGLKPLLGFDVWEHAYYLQYKNVRPDYVKAL 592 SGW WLG NK+ G L I C NQDPL+ TTGL PLLG DVWEHAYYLQYKNVRPDY+KA+ Sbjct: 145 SGWGWLGFNKERGHLQIAACPNQDPLQGTTGLIPLLGIDVWEHAYYLQYKNVRPDYLKAI 204 Query: 593 WRIINWGDVAHRF 631 W +INW +V R+ Sbjct: 205 WNVINWENVTERY 217
>sp|Q8HXP7|SODM_PANTR Superoxide dismutase [Mn], mitochondrial Length = 198 Score = 294 bits (752), Expect = 2e-79 Identities = 130/193 (67%), Positives = 155/193 (80%) Frame = +2 Query: 53 KHTLPDLPYDYNALEPSISADIMEVHHKKHHAAYVNNLNSALEQLDEAVHKNDVNKIISM 232 KH+LPDLPYDY ALEP I+A IM++HH KHHAAYVNNLN E+ EA+ K DV I++ Sbjct: 1 KHSLPDLPYDYGALEPHINAQIMQLHHSKHHAAYVNNLNVTEEKYQEALAKGDVTAQIAL 60 Query: 233 QNAIKFNGGGHLNHSIFWKNLCPKGGGEPDGSLLAALQREFGSFENFKTELSAAAVGIQG 412 Q A+KFNGGGH+NHSIFW NL P GGGEP G LL A++R+FGSF+ FK +L+AA+VG+QG Sbjct: 61 QPALKFNGGGHINHSIFWTNLSPNGGGEPKGELLEAIKRDFGSFDKFKEKLTAASVGVQG 120 Query: 413 SGWAWLGLNKKNGRLHITTCGNQDPLEATTGLKPLLGFDVWEHAYYLQYKNVRPDYVKAL 592 SGW WLG NK+ G L I C NQDPL+ TTGL PLLG DVWEHAYYLQYKNVRPDY+KA+ Sbjct: 121 SGWGWLGFNKERGHLQIAACPNQDPLQGTTGLIPLLGIDVWEHAYYLQYKNVRPDYLKAI 180 Query: 593 WRIINWGDVAHRF 631 W +INW +V R+ Sbjct: 181 WNVINWENVTERY 193
>sp|Q8HXP1|SODM_CEBAP Superoxide dismutase [Mn], mitochondrial sp|Q8HXP0|SODM_CALJA Superoxide dismutase [Mn], mitochondrial Length = 198 Score = 294 bits (752), Expect = 2e-79 Identities = 130/193 (67%), Positives = 154/193 (79%) Frame = +2 Query: 53 KHTLPDLPYDYNALEPSISADIMEVHHKKHHAAYVNNLNSALEQLDEAVHKNDVNKIISM 232 KH+LPDLPYDY ALEP I+A IM++HH KHHAAYVNNLN E+ EA+ K DV I++ Sbjct: 1 KHSLPDLPYDYGALEPHINAQIMQLHHSKHHAAYVNNLNDTEEKYKEALAKGDVTAQIAL 60 Query: 233 QNAIKFNGGGHLNHSIFWKNLCPKGGGEPDGSLLAALQREFGSFENFKTELSAAAVGIQG 412 Q A+KFNGGGH+NHSIFW NL P GGGEP G LL A++R+FGSF+ FK L+AA+VG+QG Sbjct: 61 QPALKFNGGGHINHSIFWTNLSPNGGGEPKGELLEAIKRDFGSFDKFKERLTAASVGVQG 120 Query: 413 SGWAWLGLNKKNGRLHITTCGNQDPLEATTGLKPLLGFDVWEHAYYLQYKNVRPDYVKAL 592 SGW WLG NK+ G L I C NQDPL+ TTGL PLLG DVWEHAYYLQYKNVRPDY+KA+ Sbjct: 121 SGWGWLGFNKERGHLQIAACPNQDPLQGTTGLIPLLGIDVWEHAYYLQYKNVRPDYLKAI 180 Query: 593 WRIINWGDVAHRF 631 W +INW +V R+ Sbjct: 181 WNVINWENVTERY 193
>sp|Q5FB30|SODM_MACNE Superoxide dismutase [Mn], mitochondrial precursor Length = 222 Score = 293 bits (751), Expect = 2e-79 Identities = 130/193 (67%), Positives = 156/193 (80%) Frame = +2 Query: 53 KHTLPDLPYDYNALEPSISADIMEVHHKKHHAAYVNNLNSALEQLDEAVHKNDVNKIISM 232 KH+LPDLPYDY ALEP I+A IM++HH KHHAAYVNNLN E+ EA+ K DV I++ Sbjct: 25 KHSLPDLPYDYGALEPHINAQIMQLHHSKHHAAYVNNLNVTEEKYQEALAKGDVTAQIAL 84 Query: 233 QNAIKFNGGGHLNHSIFWKNLCPKGGGEPDGSLLAALQREFGSFENFKTELSAAAVGIQG 412 Q A+KFNGGGH+NHSIFW NL P GGGEP G LL A++R+FGSFE +K +L+AA+VG+QG Sbjct: 85 QPALKFNGGGHINHSIFWTNLSPNGGGEPKGELLEAIKRDFGSFEKYKEKLTAASVGVQG 144 Query: 413 SGWAWLGLNKKNGRLHITTCGNQDPLEATTGLKPLLGFDVWEHAYYLQYKNVRPDYVKAL 592 SGW WLG NK+ G+L I C NQDPL+ TTGL PLLG DVWEHAYYLQYKNVRPDY+KA+ Sbjct: 145 SGWGWLGFNKERGQLQIAACLNQDPLQGTTGLIPLLGIDVWEHAYYLQYKNVRPDYLKAI 204 Query: 593 WRIINWGDVAHRF 631 W +INW +V R+ Sbjct: 205 WNVINWENVTERY 217
>sp|Q8HXP5|SODM_HYLLA Superoxide dismutase [Mn], mitochondrial Length = 198 Score = 292 bits (748), Expect = 5e-79 Identities = 129/193 (66%), Positives = 154/193 (79%) Frame = +2 Query: 53 KHTLPDLPYDYNALEPSISADIMEVHHKKHHAAYVNNLNSALEQLDEAVHKNDVNKIISM 232 KH+LPDLPYDY ALEP I+A IM++HH KHHAAYVNNLN E+ EA+ K DV I++ Sbjct: 1 KHSLPDLPYDYGALEPHINAQIMQLHHSKHHAAYVNNLNVTEEKYQEALAKGDVTAQIAL 60 Query: 233 QNAIKFNGGGHLNHSIFWKNLCPKGGGEPDGSLLAALQREFGSFENFKTELSAAAVGIQG 412 Q A+KFNGGGH+NHSIFW NL P GGGEP G LL A++R+FGSF+ FK +L+A +VG+QG Sbjct: 61 QPALKFNGGGHINHSIFWTNLSPNGGGEPKGELLEAIKRDFGSFDKFKEKLTATSVGVQG 120 Query: 413 SGWAWLGLNKKNGRLHITTCGNQDPLEATTGLKPLLGFDVWEHAYYLQYKNVRPDYVKAL 592 SGW WLG NK+ G L I C NQDPL+ TTGL PLLG DVWEHAYYLQYKNVRPDY+KA+ Sbjct: 121 SGWGWLGFNKERGHLQIAACPNQDPLQGTTGLIPLLGIDVWEHAYYLQYKNVRPDYLKAI 180 Query: 593 WRIINWGDVAHRF 631 W +INW +V R+ Sbjct: 181 WNVINWENVTERY 193
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 87,774,658
Number of Sequences: 369166
Number of extensions: 2004842
Number of successful extensions: 5713
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 5192
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5359
length of database: 68,354,980
effective HSP length: 107
effective length of database: 48,588,335
effective search space used: 6267895215
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail