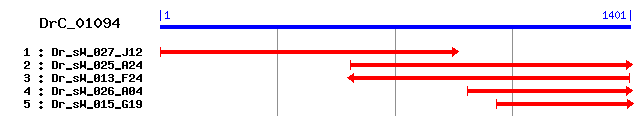
DrC_01094
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_01094 (1401 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q9ET22|DPP2_MOUSE Dipeptidyl-peptidase II precursor (DPP... 407 e-113 sp|Q9EPB1|DPP2_RAT Dipeptidyl-peptidase II precursor (DPP I... 396 e-110 sp|Q9UHL4|DPP2_HUMAN Dipeptidyl-peptidase II precursor (DPP... 385 e-106 sp|Q7TMR0|PCP_MOUSE Lysosomal Pro-X carboxypeptidase precur... 304 5e-82 sp|Q5RBU7|PCP_PONPY Lysosomal Pro-X carboxypeptidase precur... 303 8e-82 sp|P42785|PCP_HUMAN Lysosomal Pro-X carboxypeptidase precur... 302 2e-81 sp|P34676|YO26_CAEEL Putative serine protease Z688.6 precursor 248 4e-65 sp|P34610|PCP1_CAEEL Putative serine protease pcp-1 precursor 238 3e-62 sp|P34528|YM67_CAEEL Putative serine protease K12H4.7 precu... 140 1e-32 sp|Q9NQE7|TSSP_HUMAN Thymus-specific serine protease precursor 134 8e-31
>sp|Q9ET22|DPP2_MOUSE Dipeptidyl-peptidase II precursor (DPP II) (Dipeptidyl aminopeptidase II) (Quiescent cell proline dipeptidase) (Dipeptidyl peptidase 7) Length = 506 Score = 407 bits (1045), Expect = e-113 Identities = 218/447 (48%), Positives = 297/447 (66%), Gaps = 1/447 (0%) Frame = +1 Query: 1 EQILDHFNFLSVQRGYTTKFKQRFLYEDKWYKPG-GPLFFYCGNEGSIEGFWNNTGLMFE 177 EQ +DHFNF S G T F QRFL DK++K G GP+FFY GNEG I F NN+G M E Sbjct: 47 EQYMDHFNFESF--GNKT-FGQRFLVSDKFWKMGEGPIFFYTGNEGDIWSFANNSGFMVE 103 Query: 178 LAPKFNALVVFGEHRYYGVSLPFGVSKSFRQPYIQYLSVEQALADFVYMIPHVQGKLKAA 357 LA + AL+VF EHRYYG SLPFGV +S ++ Y Q L+VEQALADF ++ ++ L Sbjct: 104 LAAQQEALLVFAEHRYYGKSLPFGV-QSTQRGYTQLLTVEQALADFAVLLQALRQDLGVH 162 Query: 358 NSKVIAFGGSYGGCLAAYIRFKYPNVVLGSLAASAPIGWVSGGDGFHKFFEVITKDFREE 537 ++ IAFGGSYGG L+AY+R KYP++V G+LAASAP+ V+G ++FF +T DF + Sbjct: 163 DAPTIAFGGSYGGMLSAYMRMKYPHLVAGALAASAPVVAVAGLGDSYQFFRDVTADFYGQ 222 Query: 538 NAQCVDIVKDGFGQMRYLATQGQQGFQLISKYLKTCKPLTSISDVNYAMKWLRNGFVMLA 717 + +C V+D F Q++ L QG + IS+ TC+ L+S D+ + RN F +LA Sbjct: 223 SPKCAQAVRDAFQQIKDLFLQG--AYDTISQNFGTCQSLSSPKDLTQLFGFARNAFTVLA 280 Query: 718 MMDYPYATNFMGNLPAHPVRVACQNILANKDNSIVALREAGAVYYNTSQTSTCFSPFDLY 897 MMDYPY T+F+G LPA+PV+V CQ +L N+ I+ LR + YN+S T C+ + LY Sbjct: 281 MMDYPYPTDFLGPLPANPVKVGCQRLL-NEGQRIMGLRALAGLVYNSSGTEPCYDIYRLY 339 Query: 898 IECSDITGCGLGPDSLAWDFQACTEMKLYDDSNATSGDMFPDLPLTRQQVDKHCIKKFEV 1077 C+D TGCG G D+ AWD+QACTE+ L DSN + DMFP++P + + ++C+ + V Sbjct: 340 QSCADPTGCGTGSDARAWDYQACTEINLTFDSNNVT-DMFPEIPFSEELRQQYCLDTWGV 398 Query: 1078 DRADNQLKYLEWGDYVRTATNIIFSNGDLDPWSLGGITENASLPSSSVALWIKGGAHHLD 1257 + L+ WG ++ A+NIIFSNGDLDPW+ GGI N L +S +A+ I+GGAHHLD Sbjct: 399 WPRQDWLQTSFWGGDLKAASNIIFSNGDLDPWAGGGIQSN--LSTSVIAVTIQGGAHHLD 456 Query: 1258 LRSSNPADPPSVQQARQSEEFYIRKWL 1338 LR+SN DPPSV + R+ E IR+W+ Sbjct: 457 LRASNSEDPPSVVEVRKLESTLIREWV 483
>sp|Q9EPB1|DPP2_RAT Dipeptidyl-peptidase II precursor (DPP II) (Dipeptidyl aminopeptidase II) (Quiescent cell proline dipeptidase) (Dipeptidyl peptidase 7) Length = 500 Score = 396 bits (1018), Expect = e-110 Identities = 211/447 (47%), Positives = 292/447 (65%), Gaps = 1/447 (0%) Frame = +1 Query: 1 EQILDHFNFLSVQRGYTTKFKQRFLYEDKWYKPG-GPLFFYCGNEGSIEGFWNNTGLMFE 177 EQ +DHFNF S F QRFL DK++K G GP+FFY GNEG I NN+G + E Sbjct: 47 EQYMDHFNFESFSN---KTFGQRFLVSDKFWKMGEGPIFFYTGNEGDIWSLANNSGFIVE 103 Query: 178 LAPKFNALVVFGEHRYYGVSLPFGVSKSFRQPYIQYLSVEQALADFVYMIPHVQGKLKAA 357 LA + AL+VF EHRYYG SLPFGV +S ++ Y Q L+VEQALADF ++ ++ L Sbjct: 104 LAAQQEALLVFAEHRYYGKSLPFGV-QSTQRGYTQLLTVEQALADFAVLLQALRHNLGVQ 162 Query: 358 NSKVIAFGGSYGGCLAAYIRFKYPNVVLGSLAASAPIGWVSGGDGFHKFFEVITKDFREE 537 ++ IAFGGSYGG L+AY+R KYP++V G+LAASAP+ V+G +FF +T DF + Sbjct: 163 DAPTIAFGGSYGGMLSAYMRMKYPHLVAGALAASAPVIAVAGLGNPDQFFRDVTADFYGQ 222 Query: 538 NAQCVDIVKDGFGQMRYLATQGQQGFQLISKYLKTCKPLTSISDVNYAMKWLRNGFVMLA 717 + +C V+D F Q++ L QG + IS+ TC+ L+S D+ + RN F +LA Sbjct: 223 SPKCAQAVRDAFQQIKDLFLQG--AYDTISQNFGTCQSLSSPKDLTQLFGFARNAFTVLA 280 Query: 718 MMDYPYATNFMGNLPAHPVRVACQNILANKDNSIVALREAGAVYYNTSQTSTCFSPFDLY 897 MMDYPY TNF+G LPA+PV+V C+ +L ++ I+ LR + YN+S CF + +Y Sbjct: 281 MMDYPYPTNFLGPLPANPVKVGCERLL-SEGQRIMGLRALAGLVYNSSGMEPCFDIYQMY 339 Query: 898 IECSDITGCGLGPDSLAWDFQACTEMKLYDDSNATSGDMFPDLPLTRQQVDKHCIKKFEV 1077 C+D TGCG G ++ AWD+QACTE+ L DSN + DMFP++P + + ++C+ + V Sbjct: 340 QSCADPTGCGTGSNARAWDYQACTEINLTFDSNNVT-DMFPEIPFSDELRQQYCLDTWGV 398 Query: 1078 DRADNQLKYLEWGDYVRTATNIIFSNGDLDPWSLGGITENASLPSSSVALWIKGGAHHLD 1257 + L+ WG ++ A+NIIFSNGDLDPW+ GGI N L +S +A+ I+GGAHHLD Sbjct: 399 WPRPDWLQTSFWGGDLKAASNIIFSNGDLDPWAGGGIQRN--LSTSIIAVTIQGGAHHLD 456 Query: 1258 LRSSNPADPPSVQQARQSEEFYIRKWL 1338 LR+SN DPPSV + R+ E IR+W+ Sbjct: 457 LRASNSEDPPSVVEVRKLEATLIREWV 483
>sp|Q9UHL4|DPP2_HUMAN Dipeptidyl-peptidase II precursor (DPP II) (Dipeptidyl aminopeptidase II) (Quiescent cell proline dipeptidase) (Dipeptidyl peptidase 7) Length = 492 Score = 385 bits (990), Expect = e-106 Identities = 206/447 (46%), Positives = 288/447 (64%), Gaps = 1/447 (0%) Frame = +1 Query: 1 EQILDHFNFLSVQRGYTTKFKQRFLYEDK-WYKPGGPLFFYCGNEGSIEGFWNNTGLMFE 177 +Q LDHFNF +R F QRFL D+ W + GP+FFY GNEG + F NN+G + E Sbjct: 37 QQRLDHFNF---ERFGNKTFPQRFLVSDRFWVRGEGPIFFYTGNEGDVWAFANNSGFVAE 93 Query: 178 LAPKFNALVVFGEHRYYGVSLPFGVSKSFRQPYIQYLSVEQALADFVYMIPHVQGKLKAA 357 LA + AL+VF EHRYYG SLPFG ++S ++ + + L+VEQALADF ++ ++ L A Sbjct: 94 LAAERGALLVFAEHRYYGKSLPFG-AQSTQRGHTELLTVEQALADFAELLRALRRDLGAQ 152 Query: 358 NSKVIAFGGSYGGCLAAYIRFKYPNVVLGSLAASAPIGWVSGGDGFHKFFEVITKDFREE 537 ++ IAFGGSYGG L+AY+R KYP++V G+LAASAP+ V+G ++FF +T DF + Sbjct: 153 DAPAIAFGGSYGGMLSAYLRMKYPHLVAGALAASAPVLAVAGLGDSNQFFRDVTADFEGQ 212 Query: 538 NAQCVDIVKDGFGQMRYLATQGQQGFQLISKYLKTCKPLTSISDVNYAMKWLRNGFVMLA 717 + +C V++ F Q++ L QG + + TC+PL+ D+ + RN F +LA Sbjct: 213 SPKCTQGVREAFRQIKDLFLQG--AYDTVRWEFGTCQPLSDEKDLTQLFMFARNAFTVLA 270 Query: 718 MMDYPYATNFMGNLPAHPVRVACQNILANKDNSIVALREAGAVYYNTSQTSTCFSPFDLY 897 MMDYPY T+F+G LPA+PV+V C +L+ I LR + YN S + C+ + LY Sbjct: 271 MMDYPYPTDFLGPLPANPVKVGCDRLLSEAQR-ITGLRALAGLVYNASGSEHCYDIYRLY 329 Query: 898 IECSDITGCGLGPDSLAWDFQACTEMKLYDDSNATSGDMFPDLPLTRQQVDKHCIKKFEV 1077 C+D TGCG GPD+ AWD+QACTE+ L SN + DMFPDLP T + ++C+ + V Sbjct: 330 HSCADPTGCGTGPDARAWDYQACTEINLTFASNNVT-DMFPDLPFTDELRQRYCLDTWGV 388 Query: 1078 DRADNQLKYLEWGDYVRTATNIIFSNGDLDPWSLGGITENASLPSSSVALWIKGGAHHLD 1257 + L WG +R A+NIIFSNG+LDPW+ GGI N L +S +A+ I+GGAHHLD Sbjct: 389 WPRPDWLLTSFWGGDLRAASNIIFSNGNLDPWAGGGIRRN--LSASVIAVTIQGGAHHLD 446 Query: 1258 LRSSNPADPPSVQQARQSEEFYIRKWL 1338 LR+S+P DP SV +AR+ E I +W+ Sbjct: 447 LRASHPEDPASVVEARKLEATIIGEWV 473
>sp|Q7TMR0|PCP_MOUSE Lysosomal Pro-X carboxypeptidase precursor (Prolylcarboxypeptidase) (PRCP) (Proline carboxypeptidase) Length = 491 Score = 304 bits (778), Expect = 5e-82 Identities = 178/451 (39%), Positives = 258/451 (57%), Gaps = 5/451 (1%) Frame = +1 Query: 1 EQILDHFNFLSVQRGYTTKFKQRFLYEDK-WYKPGGPLFFYCGNEGSIEGFWNNTGLMFE 177 EQ +DHF F ++ FKQR+L DK W + GG + FY GNEG I F NNTG M++ Sbjct: 52 EQKVDHFGFADMRT-----FKQRYLVADKHWQRNGGSILFYTGNEGDIVWFCNNTGFMWD 106 Query: 178 LAPKFNALVVFGEHRYYGVSLPFGVSKSFRQPYIQYLSVEQALADFVYMIPHVQGKLKAA 357 +A + A++VF EHRYYG SLPFG ++ +L+ EQALADF +I H++ + A Sbjct: 107 VAEELKAMLVFAEHRYYGESLPFGQDSFKDSQHLNFLTSEQALADFAELIRHLEKTIPGA 166 Query: 358 NSK-VIAFGGSYGGCLAAYIRFKYPNVVLGSLAASAPIGWVSGGDGFHKFFEVITKDFRE 534 + VIA GGSYGG LAA+ R KYP++V+G+LAASAPI + G +F +++T DFR+ Sbjct: 167 QGQPVIAIGGSYGGMLAAWFRMKYPHIVVGALAASAPIWQLDGMVPCGEFMKIVTNDFRK 226 Query: 535 ENAQCVDIVKDGFGQMRYLATQGQQGFQLISKYLKTCKPLTSISDVNYAMKWLRNGFVML 714 C + ++ + + L+ G G Q ++ L C PLTS + W+ +V L Sbjct: 227 SGPYCSESIRKSWNVIDKLSGSG-SGLQSLTNILHLCSPLTS-EKIPTLKGWIAETWVNL 284 Query: 715 AMMDYPYATNFMGNLPAHPVRVACQNILANKDNSIVALR---EAGAVYYNTSQTSTCFSP 885 AM++YPYA NF+ LPA P++ CQ + + V L+ +A +VYYN S + C Sbjct: 285 AMVNYPYACNFLQPLPAWPIKEVCQYLKNPNVSDTVLLQNIFQALSVYYNYSGQAAC--- 341 Query: 886 FDLYIECSDITGCGLGPDSLAWDFQACTEMKLYDDSNATSGDMFPDLPLTRQQVDKHCIK 1065 + S T LG S+ W FQACTEM + +N DMF ++ C Sbjct: 342 ----LNISQTTTSSLG--SMGWSFQACTEMVMPFCTNGID-DMFEPFLWDLEKYSNDCFN 394 Query: 1066 KFEVDRADNQLKYLEWGDYVRTATNIIFSNGDLDPWSLGGITENASLPSSSVALWIKGGA 1245 ++ V + + + G + + +NIIFSNG+LDPWS GG+T + + + VA+ I GA Sbjct: 395 QWGVKPRPHWMTTMYGGKNISSHSNIIFSNGELDPWSGGGVTRD--ITDTLVAINIHDGA 452 Query: 1246 HHLDLRSSNPADPPSVQQARQSEEFYIRKWL 1338 HHLDLR+ N DP SV +R E +++KW+ Sbjct: 453 HHLDLRAHNAFDPSSVLLSRLLEVKHMKKWI 483
>sp|Q5RBU7|PCP_PONPY Lysosomal Pro-X carboxypeptidase precursor (Prolylcarboxypeptidase) (PRCP) (Proline carboxypeptidase) Length = 496 Score = 303 bits (776), Expect = 8e-82 Identities = 178/453 (39%), Positives = 263/453 (58%), Gaps = 5/453 (1%) Frame = +1 Query: 1 EQILDHFNFLSVQRGYTTKFKQRFLYEDK-WYKPGGPLFFYCGNEGSIEGFWNNTGLMFE 177 +Q +DHF F +V+ F QR+L DK W K GG + FY GNEG I F NNTG M++ Sbjct: 54 QQKVDHFGFNTVKT-----FNQRYLVADKYWKKNGGSILFYTGNEGDIIWFCNNTGFMWD 108 Query: 178 LAPKFNALVVFGEHRYYGVSLPFGVSKSFRQPYIQYLSVEQALADFVYMIPHVQGKLKAA 357 +A + A++VF EHRYYG SLPFG + ++ +L+ EQALADF +I H++ + A Sbjct: 109 VAEELKAMLVFAEHRYYGESLPFGDNTFKDSRHLNFLTSEQALADFAELIKHLKRTIPGA 168 Query: 358 -NSKVIAFGGSYGGCLAAYIRFKYPNVVLGSLAASAPIGWVSGGDGFHKFFEVITKDFRE 534 N VIA GGSYGG LAA+ R KYP++V+G+LAASAPI F +++T DFR+ Sbjct: 169 ENQPVIAIGGSYGGMLAAWFRMKYPHMVVGALAASAPIWQFEDLVPCGVFMKIVTTDFRK 228 Query: 535 ENAQCVDIVKDGFGQMRYLATQGQQGFQLISKYLKTCKPLTSISDVNYAMKWLRNGFVML 714 C + ++ + + L+ G G Q ++ L C PLTS D+ + W+ +V L Sbjct: 229 SGPHCSESIRRSWDAINRLSNTG-SGLQWLTGALHLCSPLTS-QDIQHLKDWISETWVNL 286 Query: 715 AMMDYPYATNFMGNLPAHPVRVACQNIL-ANKDNSIVA--LREAGAVYYNTSQTSTCFSP 885 AM+DYPYA+NF+ LPA P++V CQ + N +S++ + +A VYYN S C Sbjct: 287 AMVDYPYASNFLQPLPAWPIKVVCQYLKNPNVSDSLLLQNIFQALNVYYNYSGQVKC--- 343 Query: 886 FDLYIECSDITGCGLGPDSLAWDFQACTEMKLYDDSNATSGDMFPDLPLTRQQVDKHCIK 1065 + S+ LG +L W +QACTE+ + +N DMF +++ C + Sbjct: 344 ----LNISETATSSLG--TLGWSYQACTEVVMPFCTNGVD-DMFEPHSWNLKELSDDCFQ 396 Query: 1066 KFEVDRADNQLKYLEWGDYVRTATNIIFSNGDLDPWSLGGITENASLPSSSVALWIKGGA 1245 ++ V + + + G + + TNI+FSNG+LDPWS GG+T++ + + VA+ I GA Sbjct: 397 QWGVRPRPSWITTMYGGKNISSHTNIVFSNGELDPWSGGGVTKD--ITDTLVAVTISEGA 454 Query: 1246 HHLDLRSSNPADPPSVQQARQSEEFYIRKWLNE 1344 HHLDLR+ N DP SV AR E +++ W+ + Sbjct: 455 HHLDLRTKNALDPTSVLLARSLEVRHMKNWIRD 487
>sp|P42785|PCP_HUMAN Lysosomal Pro-X carboxypeptidase precursor (Prolylcarboxypeptidase) (PRCP) (Proline carboxypeptidase) (Angiotensinase C) (Lysosomal carboxypeptidase C) Length = 496 Score = 302 bits (773), Expect = 2e-81 Identities = 178/453 (39%), Positives = 262/453 (57%), Gaps = 5/453 (1%) Frame = +1 Query: 1 EQILDHFNFLSVQRGYTTKFKQRFLYEDK-WYKPGGPLFFYCGNEGSIEGFWNNTGLMFE 177 +Q +DHF F +V+ F QR+L DK W K GG + FY GNEG I F NNTG M++ Sbjct: 54 QQKVDHFGFNTVKT-----FNQRYLVADKYWKKNGGSILFYTGNEGDIIWFCNNTGFMWD 108 Query: 178 LAPKFNALVVFGEHRYYGVSLPFGVSKSFRQPYIQYLSVEQALADFVYMIPHVQGKLKAA 357 +A + A++VF EHRYYG SLPFG + ++ +L+ EQALADF +I H++ + A Sbjct: 109 VAEELKAMLVFAEHRYYGESLPFGDNSFKDSRHLNFLTSEQALADFAELIKHLKRTIPGA 168 Query: 358 -NSKVIAFGGSYGGCLAAYIRFKYPNVVLGSLAASAPIGWVSGGDGFHKFFEVITKDFRE 534 N VIA GGSYGG LAA+ R KYP++V+G+LAASAPI F +++T DFR+ Sbjct: 169 ENQPVIAIGGSYGGMLAAWFRMKYPHMVVGALAASAPIWQFEDLVPCGVFMKIVTTDFRK 228 Query: 535 ENAQCVDIVKDGFGQMRYLATQGQQGFQLISKYLKTCKPLTSISDVNYAMKWLRNGFVML 714 C + + + + L+ G G Q ++ L C PLTS D+ + W+ +V L Sbjct: 229 SGPHCSESIHRSWDAINRLSNTG-SGLQWLTGALHLCSPLTS-QDIQHLKDWISETWVNL 286 Query: 715 AMMDYPYATNFMGNLPAHPVRVACQNIL-ANKDNSIVA--LREAGAVYYNTSQTSTCFSP 885 AM+DYPYA+NF+ LPA P++V CQ + N +S++ + +A VYYN S C Sbjct: 287 AMVDYPYASNFLQPLPAWPIKVVCQYLKNPNVSDSLLLQNIFQALNVYYNYSGQVKC--- 343 Query: 886 FDLYIECSDITGCGLGPDSLAWDFQACTEMKLYDDSNATSGDMFPDLPLTRQQVDKHCIK 1065 + S+ LG +L W +QACTE+ + +N DMF +++ C + Sbjct: 344 ----LNISETATSSLG--TLGWSYQACTEVVMPFCTNGVD-DMFEPHSWNLKELSDDCFQ 396 Query: 1066 KFEVDRADNQLKYLEWGDYVRTATNIIFSNGDLDPWSLGGITENASLPSSSVALWIKGGA 1245 ++ V + + + G + + TNI+FSNG+LDPWS GG+T++ + + VA+ I GA Sbjct: 397 QWGVRPRPSWITTMYGGKNISSHTNIVFSNGELDPWSGGGVTKD--ITDTLVAVTISEGA 454 Query: 1246 HHLDLRSSNPADPPSVQQARQSEEFYIRKWLNE 1344 HHLDLR+ N DP SV AR E +++ W+ + Sbjct: 455 HHLDLRTKNALDPMSVLLARSLEVRHMKNWIRD 487
>sp|P34676|YO26_CAEEL Putative serine protease Z688.6 precursor Length = 507 Score = 248 bits (632), Expect = 4e-65 Identities = 161/450 (35%), Positives = 242/450 (53%), Gaps = 20/450 (4%) Frame = +1 Query: 55 KFKQRFLYEDKWYKPGGPLFFYCGNEGSIEGFWNNTGLMFELAPKFNALVVFGEHRYYGV 234 +F R+ Y+ GGP+ FY GNEGS+E F NTG M++LAP+ A VVF EHR+YG Sbjct: 60 EFDLRYFLNIDHYETGGPILFYTGNEGSLEAFAENTGFMWDLAPELKAAVVFVEHRFYGK 119 Query: 235 SLPFGVSKSFRQPYIQYLSVEQALADFVYMIPHVQG-KLKAA-NSKVIAFGGSYGGCLAA 408 S PF ++ YLS +QALADF + + K+K A S VIAFGGSYGG L+A Sbjct: 120 SQPFKNESYTDIRHLGYLSSQQALADFALSVQFFKNEKIKGAQKSAVIAFGGSYGGMLSA 179 Query: 409 YIRFKYPNVVLGSLAASAPIGWVSGGDGFHKFFE-VITKDFREENAQCVDIVKDGFGQMR 585 + R KYP++V G++AASAP+ W + + ++ ++T+ F + I K G+ + Sbjct: 180 WFRIKYPHIVDGAIAASAPVFWFTDSNIPEDVYDFIVTRAFLDAGCNRKAIEK-GWIALD 238 Query: 586 YLA--TQGQQGFQLISKYLKTCKPLTSISDVNYAMKWLRNGFVMLAMMDYPYATNFMGNL 759 LA G+Q ++ K L L + D+ + +++R +AM++YPY T+F+ +L Sbjct: 239 ELAKSDSGRQYLNVLYK-LDPKSKLENKDDIGFLKQYIRESMEAMAMVNYPYPTSFLSSL 297 Query: 760 PAHPVRVACQNIL---ANKDNSIVALREAGAVYYN-TSQTSTCFSPFDLYIECSDITGC- 924 PA PV+ AC++ ++ S L + +YYN T ST C++ C Sbjct: 298 PAWPVKEACKSASQPGKTQEESAEQLYKIVNLYYNYTGDKST---------HCANAAKCD 348 Query: 925 ---GLGPDSLAWDFQACTEMKLYDDSNATSGDMF-PDLPLTRQQVDKHCIKKF-EVDRAD 1089 G D L W FQ CTEM + + D F D P T ++ + C++ F + Sbjct: 349 SAYGSLGDPLGWPFQTCTEMVMPLCGSGYPNDFFWKDCPFTSEKYAEFCMQTFSSIHYNK 408 Query: 1090 NQLKYLEWG-----DYVRTATNIIFSNGDLDPWSLGGITENASLPSSSVALWIKGGAHHL 1254 L+ L G + +A+NI+FSNG LDPWS GG + + S +++ +K GAHH Sbjct: 409 TLLRPLAGGLAFGATSLPSASNIVFSNGYLDPWSGGGYDHSDKVQGSVISVILKQGAHHY 468 Query: 1255 DLRSSNPADPPSVQQARQSEEFYIRKWLNE 1344 DLR ++P D V++ R E I+KW+ E Sbjct: 469 DLRGAHPQDTEEVKKVRAMETQAIKKWIKE 498
>sp|P34610|PCP1_CAEEL Putative serine protease pcp-1 precursor Length = 565 Score = 238 bits (607), Expect = 3e-62 Identities = 164/468 (35%), Positives = 243/468 (51%), Gaps = 22/468 (4%) Frame = +1 Query: 10 LDHFNFLSVQRGYTTKFKQRFLYEDKWYKPGGPLFFYCGNEGSIEGFWNNTGLMFELAPK 189 LDHF + G T F R ++ + +YKPGGP+FFY GNEG +E F TG+MF+LAP Sbjct: 51 LDHFTW-----GDTRTFDMRVMWNNTFYKPGGPIFFYTGNEGGLESFVTATGMMFDLAPM 105 Query: 190 FNALVVFGEHRYYGVSLPFGVSKSFRQPYIQYLSVEQALADFVYMIPHV-----QGKLK- 351 FNA ++F EHR+YG + PFG + YL+ EQALAD+ ++ + Q K+ Sbjct: 106 FNASIIFAEHRFYGQTQPFGNQSYASLANVGYLTSEQALADYAELLTELKRDNNQFKMTF 165 Query: 352 AANSKVIAFGGSYGGCLAAYIRFKYPNVVLGSLAASAPIGWVSGGDGFHKFFEVITKDFR 531 A ++VI+FGGSYGG L+A+ R KYP++V G+ A SAP+ +++GG F+ IT Sbjct: 166 PAATQVISFGGSYGGMLSAWFRQKYPHIVKGAWAGSAPLIYMNGGGVDPGAFDHITSRTY 225 Query: 532 EENAQCVDIVKDGFGQMRYLATQGQQGFQLISKYLKTCKPLTSI---SDVNYAMKWLRNG 702 +N I+ + + L++ L + + P T I +D +LR Sbjct: 226 IDNGCNRFILANAWNATLNLSSTDAGRQWLNNNTVFKLDPRTKIRNQTDGWNLNAYLREA 285 Query: 703 FVMLAMMDYPYATNFMGNLPAHPVRVACQNILAN----KDNSIV-ALREAGAVYYNTSQT 867 +AM+DYPY T F+ LPA PV VAC + AN D +V A+ A +YYN ++ Sbjct: 286 IEYMAMVDYPYPTGFLEPLPAWPVTVACGYMNANGTSFSDKDLVKAVANAANIYYNYNRD 345 Query: 868 STCFSPFDLYIECSDITGCGLGPDSLAWDFQACTEMKLYDDSNATSGDMF-----PDLPL 1032 D I C D GLG D L W +Q C+E+ + ++ S D+F D+ Sbjct: 346 PNFTYCIDFSI-CGDQGTGGLGGDELGWPWQECSEIIMAMCASGGSNDVFWNECGKDIYQ 404 Query: 1033 TRQQVDKHCIKKFEVDRADNQLKYLE--WGDYVRTATNIIFSNGDLDPWSLGGITENASL 1206 T QQ K + + ++ +G + ++N+I + G LDPWS GG + + Sbjct: 405 TLQQGCVSIFKSMGWTPKNWNIDAVKTLYGYDLSGSSNLILTQGHLDPWSGGGYKVDQNN 464 Query: 1207 PSSSV-ALWIKGGAHHLDLRSSNPADPPSVQQARQSEEFYIRKWLNEN 1347 + + L I G AHHLDLR N DP +V AR ++ W++ N Sbjct: 465 AARGIYVLEIPGSAHHLDLRQPNTCDPNTVTNARFQIIQILKCWVDPN 512
>sp|P34528|YM67_CAEEL Putative serine protease K12H4.7 precursor Length = 510 Score = 140 bits (352), Expect = 1e-32 Identities = 131/465 (28%), Positives = 206/465 (44%), Gaps = 17/465 (3%) Frame = +1 Query: 4 QILDHFNFLSVQRGYTTKFKQRFLYEDKWYKPGGPLFFYCGNEGSIEGFW-NNTGL-MFE 177 Q LDHF+ SV + F+QR+ + ++WYK GGP F G EG +W + GL + Sbjct: 65 QTLDHFDS-SVGK----TFQQRYYHNNQWYKAGGPAFLMLGGEGPESSYWVSYPGLEITN 119 Query: 178 LAPKFNALVVFGEHRYYGVSLPFGVSKSFRQPYIQYLSVEQALADFVYMIPHVQGKL-KA 354 LA K A V EHR+YG + P + P ++YLS QA+ D I + K + Sbjct: 120 LAAKQGAWVFDIEHRFYGETHP---TSDMSVPNLKYLSSAQAIEDAAAFIKAMTAKFPQL 176 Query: 355 ANSKVIAFGGSYGGCLAAYIRFKYPNVVLGSLAASAPIGWVSGGDGFHKFFEVITKDFRE 534 AN+K + FGGSY G LAA+ R K+P +V ++ +S P V F ++ EV+ Sbjct: 177 ANAKWVTFGGSYSGALAAWTRAKHPELVYAAVGSSGP---VQAEVDFKEYLEVVQNSITR 233 Query: 535 ENAQCVDIVKDGFGQMRYLATQGQQGFQLISKYLKTCKPL-TSISDVNYAMKWLRNGFVM 711 + +C V GF + L Q G + + C+ + + Y + + + ++ Sbjct: 234 NSTECAASVTQGFNLVASL-LQTSDGRKQLKTAFHLCQDIQMDDKSLKYFWETVYSPYME 292 Query: 712 LAMMDYPYATNFMGNLP-AHPVRVACQNILANKDNSIVALREAGAVYYNTSQTSTCFS-P 885 + A +F L +H + C+ + K + L++ + S C Sbjct: 293 VVQYSGDAAGSFATQLTISHAI---CRYHINTKSTPLQKLKQVNDYFNQVSGYFGCNDID 349 Query: 886 FDLYIECSDITGCGLGPDSLAWDFQACTEMKLYDD-SNATSGDMFPDLP-LTRQQVDKHC 1059 ++ +I G AW +Q CTE Y S+AT+G F + L Q C Sbjct: 350 YNGFISFMKDETFGEAQSDRAWVWQTCTEFGYYQSTSSATAGPWFGGVSNLPAQYYIDEC 409 Query: 1060 IKKF-------EVDRA-DNQLKYLEWGDYVRTATNIIFSNGDLDPW-SLGGITENASLPS 1212 + EV + D +Y D + T I+ NGD+DPW +LG +T S S Sbjct: 410 TAIYGAAYNSQEVQTSVDYTNQYYGGRDNLNT-DRILLPNGDIDPWHALGKLT---SSNS 465 Query: 1213 SSVALWIKGGAHHLDLRSSNPADPPSVQQARQSEEFYIRKWLNEN 1347 + V + I G AH D+ ++ D + ARQ + WL+ N Sbjct: 466 NIVPVVINGTAHCADMYGASSLDSMYLTNARQRISDVLDGWLHAN 510
>sp|Q9NQE7|TSSP_HUMAN Thymus-specific serine protease precursor Length = 514 Score = 134 bits (336), Expect = 8e-31 Identities = 134/467 (28%), Positives = 204/467 (43%), Gaps = 21/467 (4%) Frame = +1 Query: 1 EQILDHFNFLSVQRGYTTKFKQRFLYEDK-WYKPGGPLFFYCGNEGSIEGFWNNTGLMFE 177 EQ+LD FN +S +R F QR+ D+ W GP+F + G EGS+ G Sbjct: 63 EQLLDPFN-VSDRRS----FLQRYWVNDQHWVGQDGPIFLHLGGEGSLGPGSVMRGHPAA 117 Query: 178 LAPKFNALVVFGEHRYYGVSLPFGVSKSFRQPYIQYLSVEQALADFVYMIPHVQGKLKAA 357 LAP + ALV+ EHR+YG+S+P G +++LS ALAD V + + Sbjct: 118 LAPAWGALVISLEHRFYGLSIPAG---GLEMAQLRFLSSRLALADVVSARLALSRLFNIS 174 Query: 358 NSKV-IAFGGSYGGCLAAYIRFKYPNVVLGSLAASAPIGWVSGGDGFHKFFEVITKDFRE 534 +S I FGGSY G LAA+ R K+P+++ S+A+SAP+ V F ++ +V+++ Sbjct: 175 SSSPWICFGGSYAGSLAAWARLKFPHLIFASVASSAPVRAVL---DFSEYNDVVSRSLMS 231 Query: 535 E----NAQCVDIVKDGFGQMRYLATQGQQGFQLISKYLKTCKPL---TSISDVNYAMKWL 693 + +C V F ++ G + L C PL + +++ A++ L Sbjct: 232 TAIGGSLECRAAVSVAFAEVERRLRSGGAAQAALRTELSACGPLGRAENQAELLGALQAL 291 Query: 694 RNGFVMLAMMDYPYATNFMGNLPAH-PVRVACQNILANKDN-----SIVALREA-GAVYY 852 G V + G A VR C +L N LR A V + Sbjct: 292 VGGVV-----------QYDGQTGAPLSVRQLCGLLLGGGGNRSHSTPYCGLRRAVQIVLH 340 Query: 853 NTSQTSTCFSPFDLYIECSDITGCGLGPDSLAWDFQACTEMKLYDDSNATSGDMFPDLPL 1032 + Q FS + + G W +Q CTE Y F LP Sbjct: 341 SLGQKCLSFSRAETVAQLRSTEPQLSGVGDRQWLYQTCTEFGFYVTCE-NPRCPFSQLPA 399 Query: 1033 TRQQVDKHCIKKF-----EVDRADNQLKYLEWGDYVRTATNIIFSNGDLDPWSLGGITEN 1197 Q+D C + F V +A Q +G A ++F NGD DPW + +T+ Sbjct: 400 LPSQLDL-CEQVFGLSALSVAQAVAQTNSY-YGGQTPGANKVLFVNGDTDPWHVLSVTQ- 456 Query: 1198 ASLPSSSVALWIKGGAHHLDLRSSNPADPPSVQQARQSEEFYIRKWL 1338 +L SS L I+ G+H LD+ P+D PS++ RQ+ ++ WL Sbjct: 457 -ALGSSESTLLIRTGSHCLDMAPERPSDSPSLRLGRQNIFQQLQTWL 502
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 170,115,731
Number of Sequences: 369166
Number of extensions: 3797732
Number of successful extensions: 9303
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 8742
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 9237
length of database: 68,354,980
effective HSP length: 114
effective length of database: 47,295,190
effective search space used: 16647906880
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail