DrC_01000
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_01000 (1096 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q9U4L6|TOM40_DROME Probable mitochondrial import recepto... 207 4e-53 sp|Q9QYA2|TOM40_MOUSE Probable mitochondrial import recepto... 178 2e-44 sp|O96008|TOM40_HUMAN Probable mitochondrial import recepto... 177 5e-44 sp|Q18090|TOM40_CAEEL Probable mitochondrial import recepto... 171 4e-42 sp|O13656|TOM40_SCHPO Probable mitochondrial import recepto... 76 1e-13 sp|P23644|TOM40_YEAST Mitochondrial import receptor subunit... 55 4e-07 sp|Q9LHE5|TOM40_ARATH Probable mitochondrial import recepto... 40 0.012 sp|Q59088|G6PI_ACIAD Glucose-6-phosphate isomerase (GPI) (P... 32 3.2 sp|P19812|UBR1_YEAST Ubiquitin-protein ligase E3 component ... 31 5.5 sp|O74161|CHS5_CANAL Chitin biosynthesis protein CHS5 31 5.5
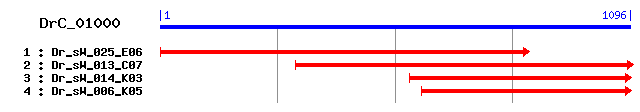
>sp|Q9U4L6|TOM40_DROME Probable mitochondrial import receptor subunit TOM40 homolog (Translocase of outer membrane 40 kDa subunit homolog) (Male sterile protein 15) Length = 344 Score = 207 bits (527), Expect = 4e-53 Identities = 101/287 (35%), Positives = 167/287 (58%), Gaps = 2/287 (0%) Frame = +1 Query: 184 TMESLHNITHEIFPVPFEGVKLVVNQGFNQHFQLNHSFAFGSEKQSGYKFGATYVGNTKL 363 T+E LH +I + FEG K+++N+G + HFQ++H+ + SGY+FGATYVG + Sbjct: 62 TVEELHKKCKDIQAITFEGAKIMLNKGLSNHFQVSHTINMSNVVPSGYRFGATYVGTKEF 121 Query: 364 SETEIYPIMLGELSPNGNLNAQIIHQFNQKIKCKYVSQMKVGKIDGQQFSIEHRNKNINL 543 S TE +P++LG++ P GNLNA +IHQF+ +++CK+ SQ++ K+ Q + ++R + L Sbjct: 122 SPTEAFPVLLGDIDPAGNLNANVIHQFSARLRCKFASQIQESKVVASQLTTDYRGSDYTL 181 Query: 544 GFSIVNPSIQNKEISCIFDVMRRITDKLSLGSMFVFQRSNQAQFTNFGVWNVGGQYVGNK 723 ++ NPSI + ++ +T L+LGS +Q + +V G+Y Sbjct: 182 SLTVANPSIFTNSGVVVGQYLQSVTPALALGSELAYQFGPNVPGRQIAIMSVVGRYTAGS 241 Query: 724 --WTAAVGVRPFQSSLHSTFHMKLNEAFQIGTEIETSLGHRQCVGTAGYQFDFPKSGITI 897 W+ +G QS LH ++ K ++ QIG E+ETSL ++ V T YQ D PK+ + Sbjct: 242 SVWSGTLG----QSGLHVCYYQKASDQLQIGAEVETSLRMQESVATLAYQIDLPKANLVF 297 Query: 898 KSQVDSNWKIATSYERKFIAPIPFSINLCINGDIWKSNYGLGVGFSI 1038 + +DSNW+I E++ +AP+PF++ L + K+N+ LG G I Sbjct: 298 RGGIDSNWQIFGVLEKR-LAPLPFTLALSGRMNHVKNNFRLGCGLMI 343
>sp|Q9QYA2|TOM40_MOUSE Probable mitochondrial import receptor subunit TOM40 homolog (Translocase of outer membrane 40 kDa subunit homolog) Length = 359 Score = 178 bits (452), Expect = 2e-44 Identities = 95/285 (33%), Positives = 155/285 (54%) Frame = +1 Query: 184 TMESLHNITHEIFPVPFEGVKLVVNQGFNQHFQLNHSFAFGSEKQSGYKFGATYVGNTKL 363 T E H E+FPV EGVKL VN+G + FQ+ H+ A G+ +S Y FG TYVG +L Sbjct: 80 TFEECHRKCKELFPVQMEGVKLTVNKGLSNRFQVTHTVALGTIGESNYHFGVTYVGTKQL 139 Query: 364 SETEIYPIMLGELSPNGNLNAQIIHQFNQKIKCKYVSQMKVGKIDGQQFSIEHRNKNINL 543 S TE +P+++G++ +G+LNAQ+IHQ + ++ K Q + K Q E+R + Sbjct: 140 SPTEAFPVLVGDMDNSGSLNAQVIHQLSPGLRSKMAIQTQQSKFVNWQVDGEYRGSDFTA 199 Query: 544 GFSIVNPSIQNKEISCIFDVMRRITDKLSLGSMFVFQRSNQAQFTNFGVWNVGGQYVGNK 723 ++ NP + + ++ IT L+LG V+ R + T V ++ G+Y N Sbjct: 200 AVTLGNPDVLVGSGILVAHYLQSITPCLALGGELVYHRRPGEEGT---VMSLAGKYTLNN 256 Query: 724 WTAAVGVRPFQSSLHSTFHMKLNEAFQIGTEIETSLGHRQCVGTAGYQFDFPKSGITIKS 903 W A V + Q+ +H+T++ K ++ Q+G E E S + + GYQ D PK+ K Sbjct: 257 WLATVTLG--QAGMHATYYHKASDQLQVGVEFEASTRMQDTSASFGYQLDLPKANFLFKG 314 Query: 904 QVDSNWKIATSYERKFIAPIPFSINLCINGDIWKSNYGLGVGFSI 1038 V+SNW + + E+K + P+P +++LC + K+ + G G +I Sbjct: 315 SVNSNWIVGATLEKK-LPPLPLTLSLCAFLNHRKNKFLCGFGLTI 358
>sp|O96008|TOM40_HUMAN Probable mitochondrial import receptor subunit TOM40 homolog (Translocase of outer membrane 40 kDa subunit homolog) (Haymaker protein) (p38.5) Length = 361 Score = 177 bits (449), Expect = 5e-44 Identities = 95/285 (33%), Positives = 154/285 (54%) Frame = +1 Query: 184 TMESLHNITHEIFPVPFEGVKLVVNQGFNQHFQLNHSFAFGSEKQSGYKFGATYVGNTKL 363 T E H E+FP+ EGVKL VN+G + HFQ+NH+ A + +S Y FG TYVG +L Sbjct: 82 TFEECHRKCKELFPIQMEGVKLTVNKGLSNHFQVNHTVALSTIGESNYHFGVTYVGTKQL 141 Query: 364 SETEIYPIMLGELSPNGNLNAQIIHQFNQKIKCKYVSQMKVGKIDGQQFSIEHRNKNINL 543 S TE +P+++G++ +G+LNAQ+IHQ ++ K Q + K Q E+R + Sbjct: 142 SPTEAFPVLVGDMDNSGSLNAQVIHQLGPGLRSKMAIQTQQSKFVNWQVDGEYRGSDFTA 201 Query: 544 GFSIVNPSIQNKEISCIFDVMRRITDKLSLGSMFVFQRSNQAQFTNFGVWNVGGQYVGNK 723 ++ NP + + ++ IT L+LG V+ R + T V ++ G+Y N Sbjct: 202 AVTLGNPDVLVGSGILVAHYLQSITPCLALGGELVYHRRPGEEGT---VMSLAGKYTLNN 258 Query: 724 WTAAVGVRPFQSSLHSTFHMKLNEAFQIGTEIETSLGHRQCVGTAGYQFDFPKSGITIKS 903 W A V + Q+ +H+T++ K ++ Q+G E E S + + GYQ D PK+ + K Sbjct: 259 WLATVTLG--QAGMHATYYHKASDQLQVGVEFEASTRMQDTSVSFGYQLDLPKANLLFKG 316 Query: 904 QVDSNWKIATSYERKFIAPIPFSINLCINGDIWKSNYGLGVGFSI 1038 VDSNW + + E+K + P+P ++ L + K+ + G G +I Sbjct: 317 SVDSNWIVGATLEKK-LPPLPLTLALGAFLNHRKNKFQCGFGLTI 360
>sp|Q18090|TOM40_CAEEL Probable mitochondrial import receptor subunit TOM40 homolog (Translocase of outer membrane 40 kDa subunit homolog) Length = 301 Score = 171 bits (432), Expect = 4e-42 Identities = 91/283 (32%), Positives = 151/283 (53%) Frame = +1 Query: 190 ESLHNITHEIFPVPFEGVKLVVNQGFNQHFQLNHSFAFGSEKQSGYKFGATYVGNTKLSE 369 E LH ++FP FEG KL+VN+G + HFQ++H+ + S +GY+FGATYVG ++ Sbjct: 22 EELHRKARDVFPTCFEGAKLMVNKGLSSHFQVSHTLSL-SAMNTGYRFGATYVGTNQVGP 80 Query: 370 TEIYPIMLGELSPNGNLNAQIIHQFNQKIKCKYVSQMKVGKIDGQQFSIEHRNKNINLGF 549 E YPI+LG+ NGN A I+HQ + K Q++ GK+ G Q +IE + + LG Sbjct: 81 AEAYPILLGDTDVNGNTTATILHQLGI-YRTKLQGQIQQGKLAGAQATIERKGRLSTLGL 139 Query: 550 SIVNPSIQNKEISCIFDVMRRITDKLSLGSMFVFQRSNQAQFTNFGVWNVGGQYVGNKWT 729 ++ N + N+ + +RR+T +L +G+ V+Q V + +Y N + Sbjct: 140 TLANIDLVNEAGILVGQFLRRLTPRLDVGTEMVYQYGKNIPGGQISVLSYAARYTANHFI 199 Query: 730 AAVGVRPFQSSLHSTFHMKLNEAFQIGTEIETSLGHRQCVGTAGYQFDFPKSGITIKSQV 909 AA + S +H T++ K NE G E E + + V T YQ + P+ G+T+++ Sbjct: 200 AAATLGA--SGVHLTYYHKQNENLAFGVEFECNANVGEAVTTLAYQTELPEEGVTMRASF 257 Query: 910 DSNWKIATSYERKFIAPIPFSINLCINGDIWKSNYGLGVGFSI 1038 D+NW + +E++ +PF++ L + K+ G+G I Sbjct: 258 DTNWTVGGVFEKRLSQQLPFTLALSGTLNHVKAAGKFGIGLII 300
>sp|O13656|TOM40_SCHPO Probable mitochondrial import receptor subunit tom40 (Translocase of outer membrane 40 kDa subunit) Length = 344 Score = 76.3 bits (186), Expect = 1e-13 Identities = 66/286 (23%), Positives = 122/286 (42%), Gaps = 17/286 (5%) Frame = +1 Query: 232 FEGVKLVVNQGF--NQHFQLNHSFAFGSEKQSGYKFGATYVGNTKLSETEIYPIML-GEL 402 F GV+ V +GF + F ++H+FA GS+ Y F + G P+ L G + Sbjct: 51 FTGVRADVTKGFCTSPWFTVSHAFALGSQVLPPYSFSTMFGGE---------PLFLRGSV 101 Query: 403 SPNGNLNAQIIHQFNQKIKCKYVSQMKVGKIDGQ-QFSIEHRNKNINLGFSIVNPSIQNK 579 +G + A + +N + K Q+ G + Q +H+ K+ + F +NP + K Sbjct: 102 DNDGAVQAMLNCTWNSNVLSKVQMQLSNGAVPNMCQIEHDHKGKDFSFSFKAMNPWYEEK 161 Query: 580 EISC-IFDVMRRITDKLSLGSMFVFQRSNQAQFTNFGVWNVGGQYVGNKWTAAVGVRPFQ 756 I +++ +T KLSLG ++Q+ + + + +Y W A + Q Sbjct: 162 LTGIYIISLLQSVTPKLSLGVEALWQKPSSSIGPEEATLSYMTRYNAADWIATAHLNGSQ 221 Query: 757 SSLHSTFHMKLNEAFQIGTEIETS---LGHRQCVGT---------AGYQFDFPKSGITIK 900 + +TF KL+ + G E + S L H + T G +++F +S + Sbjct: 222 GDVTATFWRKLSPKVEAGVECQLSPVGLNHSAALMTGPKPEGLTSVGVKYEFAQS--IYR 279 Query: 901 SQVDSNWKIATSYERKFIAPIPFSINLCINGDIWKSNYGLGVGFSI 1038 QVDS ++ ER+ I + + ++ + GLG+ + Sbjct: 280 GQVDSKGRVGVYLERRLAPAITLAFSSELDHPNRNAKVGLGLSLEL 325
>sp|P23644|TOM40_YEAST Mitochondrial import receptor subunit TOM40 (Mitochondrial import-site-protein ISP42) (Translocase of outer membrane 40 kDa subunit) Length = 387 Score = 55.1 bits (131), Expect = 4e-07 Identities = 69/307 (22%), Positives = 138/307 (44%), Gaps = 25/307 (8%) Frame = +1 Query: 184 TMESLHN-ITHEIFPVP--FEGVKLVVNQGF--NQHFQLNHSFAFGSEKQSGYKFGATYV 348 T+E+L+ ++ ++F F G++ +N+ F N FQ +H+F+ GS+ Y F A + Sbjct: 62 TVENLNKEVSRDVFLSQYFFTGLRADLNKAFSMNPAFQTSHTFSIGSQALPKYAFSALFA 121 Query: 349 GNTKLSETEIYPIMLGELSPNGNLNAQIIHQFNQKIKCKYVSQMKVGKIDGQQFSIEHRN 528 + ++ I +LS +G LN + +++K K Q+ G+ Q +++ Sbjct: 122 NDNLFAQGNID----NDLSVSGRLN----YGWDKKNISKVNLQISDGQPTMCQLEQDYQA 173 Query: 529 KNINLGFSIVNPSIQNK-EIS--CIFDVMRRITDKLSLGSMFVFQRSNQAQFTNFGV--- 690 + ++ +NPS K E + + ++ +T +L+LG ++ R++ + + GV Sbjct: 174 SDFSVNVKTLNPSFSEKGEFTGVAVASFLQSVTPQLALGLETLYSRTDGSAPGDAGVSYL 233 Query: 691 ---------WNVGGQYVGNKWTAAVGVRPFQSSLHSTFHMKLNEAFQIGTE--IETSLGH 837 W GQ N A R ++ + L T+ + T +G Sbjct: 234 TRYVSKKQDWIFSGQLQANGALIASLWRKVAQNVEAGIETTLQAGMVPITDPLMGTPIGI 293 Query: 838 RQCV---GTAGYQFDFPKSGITIKSQVDSNWKIATSYERKFIAPIPFSINLCINGDIWKS 1008 + V T G ++++ +S + +DSN K+A ERK + + S+ C D +K+ Sbjct: 294 QPTVEGSTTIGAKYEYRQS--VYRGTLDSNGKVACFLERKVLPTL--SVLFCGEIDHFKN 349 Query: 1009 NYGLGVG 1029 + +G G Sbjct: 350 DTKIGCG 356
>sp|Q9LHE5|TOM40_ARATH Probable mitochondrial import receptor subunit TOM40 homolog (Translocase of outer membrane 40 kDa subunit homolog) Length = 309 Score = 40.0 bits (92), Expect = 0.012 Identities = 63/318 (19%), Positives = 124/318 (38%), Gaps = 21/318 (6%) Frame = +1 Query: 148 KMAESSDSKGLPT---MESLHN-ITHEIFPVPFEGVKLVVNQGFNQHFQLNHSFAFGSEK 315 K+ E D LP+ E LH + FEG++ + NQ F L+HS G + Sbjct: 18 KVDEKVDYSNLPSPVPYEELHREALMSLKSDNFEGLRFDFTRALNQKFSLSHSVMMGPTE 77 Query: 316 -------------QSGYKFGATYVGNTKLSETEIYPIMLGELSPNGNLNAQIIHQFNQKI 456 + Y+FGA Y + KL +++G + +G LNA++ K+ Sbjct: 78 VPAQSPETTIKIPTAHYEFGANYY-DPKL-------LLIGRVMTDGRLNARLKADLTDKL 129 Query: 457 KCKYVSQMKVGKIDGQQFSIEHRNK---NINLGFSIVNPSIQNKEISCI-FDVMRRITDK 624 V K + + EH ++ N + S +Q + + I ++ +T+ Sbjct: 130 ---------VVKANALITNEEHMSQAMFNFDYMGSDYRAQLQLGQSALIGATYIQSVTNH 180 Query: 625 LSLGSMFVFQRSNQAQFTNFGVWNVGGQYVGNKWTAAVGVRPFQSSLHSTFHMKLNEAFQ 804 LSLG + + + + V + A+ G ++ + K+++ Sbjct: 181 LSLGGEIFWAGVPRKSGIGYAARYETDKMVASGQVASTG------AVVMNYVQKISDKVS 234 Query: 805 IGTEIETSLGHRQCVGTAGYQFDFPKSGITIKSQVDSNWKIATSYERKFIAPIPFSINLC 984 + T+ + R + GY D+ ++ ++DSN + E + + F ++ Sbjct: 235 LATDFMYNYFSRDVTASVGY--DYMLRQARVRGKIDSNGVASALLEERLSMGLNFLLSAE 292 Query: 985 INGDIWKSNYGLGVGFSI 1038 + D K +Y G G ++ Sbjct: 293 L--DHKKKDYKFGFGLTV 308
>sp|Q59088|G6PI_ACIAD Glucose-6-phosphate isomerase (GPI) (Phosphoglucose isomerase) (PGI) (Phosphohexose isomerase) (PHI) Length = 557 Score = 32.0 bits (71), Expect = 3.2 Identities = 15/44 (34%), Positives = 25/44 (56%), Gaps = 1/44 (2%) Frame = +1 Query: 352 NTKLSETEIYPIMLGELSPNG-NLNAQIIHQFNQKIKCKYVSQM 480 N + E PI+ GE+ PN + Q++HQ QK+ C +++ M Sbjct: 370 NGQKVENTTCPIVWGEVGPNAQHAFYQLLHQGTQKVSCDFIAPM 413
>sp|P19812|UBR1_YEAST Ubiquitin-protein ligase E3 component N-recognin-1 (N-end-recognizing protein) Length = 1950 Score = 31.2 bits (69), Expect = 5.5 Identities = 17/71 (23%), Positives = 35/71 (49%) Frame = -2 Query: 888 TRFREIKLITSCTHALTMTQTSLNFCTYLKCFIQLHVKS*M*RTLERPNSHCRSPFITHI 709 T+ +EIKL + + L+F L C +++H+++ + N +C PF + Sbjct: 1676 TQSKEIKLREERSQHMKNADNRLDFKICLTCGVKVHLRADRHEMTKHLNKNCFKPFGAFL 1735 Query: 708 LPTNIPYTEIC 676 +P + +E+C Sbjct: 1736 MPNS---SEVC 1743
>sp|O74161|CHS5_CANAL Chitin biosynthesis protein CHS5 Length = 562 Score = 31.2 bits (69), Expect = 5.5 Identities = 17/58 (29%), Positives = 29/58 (50%) Frame = +1 Query: 358 KLSETEIYPIMLGELSPNGNLNAQIIHQFNQKIKCKYVSQMKVGKIDGQQFSIEHRNK 531 K+++ + LG+L+PN N + I + + + KY Q +V K+D F NK Sbjct: 165 KMTDLSGITVCLGDLTPNDQFNKEDIEEALKNMGAKYPVQQQV-KVDTTHFLCTRENK 221
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 127,561,315
Number of Sequences: 369166
Number of extensions: 2680870
Number of successful extensions: 5593
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 5330
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5580
length of database: 68,354,980
effective HSP length: 112
effective length of database: 47,664,660
effective search space used: 12011494320
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail