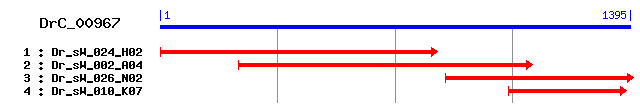
DrC_00967
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00967 (1395 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q9WUB0|UB7I3_MOUSE Ubiquitin-conjugating enzyme 7-intera... 105 2e-22 sp|Q9BYM8|UB7I3_HUMAN Ubiquitin-conjugating enzyme 7-intera... 105 4e-22 sp|Q62921|UB7I3_RAT Ubiquitin-conjugating enzyme 7-interact... 105 4e-22 sp|Q924T7|RNF31_MOUSE RING finger protein 31 59 4e-08 sp|Q96EP0|RNF31_HUMAN RING finger protein 31 (Zinc in-betwe... 57 2e-07 sp|Q8IWT3|PARC_HUMAN p53-associated parkin-like cytoplasmic... 55 5e-07 sp|Q80TT8|PARC_MOUSE p53-associated parkin-like cytoplasmic... 55 6e-07 sp|Q925F3|UB7I4_MOUSE Ubiquitin-conjugating enzyme 7-intera... 52 5e-06 sp|P58283|UB7I1_MOUSE E3 ubiquitin ligase TRIAD3 (Ubiquitin... 50 2e-05 sp|Q9NWF9|UB7I1_HUMAN E3 ubiquitin ligase TRIAD3 (Ubiquitin... 50 2e-05
>sp|Q9WUB0|UB7I3_MOUSE Ubiquitin-conjugating enzyme 7-interacting protein 3 (UbcM4-interacting protein 28) Length = 498 Score = 105 bits (263), Expect = 2e-22 Identities = 69/244 (28%), Positives = 113/244 (46%), Gaps = 29/244 (11%) Frame = +3 Query: 516 QMKNNAYIPNSESFECGICNSFILpreGLIIRNCFHCYCKSCFEETLKNVN--------F 671 Q++ + + N+E EC +C S + P E +++R C H +C+ C + T++N Sbjct: 255 QLEQRSLVLNTEPTECPVCYSVLAPGEAVVLRECLHTFCRECLQGTIRNSQEAEVACPFI 314 Query: 672 TSNFKCPKCGIEINNQEIIKAVGVEKLIGMACKEAAPK---MFCCKYPDCDG--MWDNDV 836 S + CP +E + ++ ++ + + A + + CK PDC G +++DV Sbjct: 315 DSTYSCPGKLLEREIRALLSPEDYQRFLDLGVSIAENRSTLSYHCKTPDCRGWCFFEDDV 374 Query: 837 IRKGSFICPVCKHSNCLDCEDVHEDKSCEEFR-----LTERYIDAQKA----KKKLQGES 989 F CPVC NCL C+ +HE +C E++ + + A++ K LQ Sbjct: 375 ---NEFTCPVCTRVNCLLCKAIHEHMNCREYQDDLALRAQNDVAARQTTEMLKVMLQQGE 431 Query: 990 GIKCICGRFLVQ-KGGCGALHCDSCGRQICVFTKKIA-GNNGP-DTCGCTPTKT----CH 1148 + C R +VQ K GC + C C +IC TK G GP DT G + CH Sbjct: 432 AMHCPQCRIVVQKKDGCDWIRCTVCHTEICWVTKGPRWGPGGPGDTSGGCRCRVNGIPCH 491 Query: 1149 PDCK 1160 P C+ Sbjct: 492 PSCQ 495
Score = 64.7 bits (156), Expect = 6e-10
Identities = 48/202 (23%), Positives = 81/202 (40%), Gaps = 8/202 (3%)
Frame = +3
Query: 63 CMKCKGTGIVDDSFTLRLCNHMFCHKCLCEYIKFHINCKIKCP----VENCNKNISDQII 230
C C ++ LR C H FC +CL I+ ++ CP +C + ++ I
Sbjct: 270 CPVCYSVLAPGEAVVLRECLHTFCRECLQGTIRNSQEAEVACPFIDSTYSCPGKLLEREI 329
Query: 231 RKCLSNPEIALYEQISLC----KSTKSARKCLTSNCSAIMFIEDNKAIFQCQFCDQPNCL 398
R LS + + + + +ST S C T +C F ED+ F C C + NCL
Sbjct: 330 RALLSPEDYQRFLDLGVSIAENRSTLSYH-CKTPDCRGWCFFEDDVNEFTCPVCTRVNCL 388
Query: 399 KCEKRHYPNQCGDLNHNEDAIEAAAKVAMSRHRREALEEQMKNNAYIPNSESFECGICNS 578
C+ H C + + +D A +R E L+ ++ E+ C C
Sbjct: 389 LCKAIHEHMNCRE--YQDDLALRAQNDVAARQTTEMLKVMLQ------QGEAMHCPQCRI 440
Query: 579 FILpreGLIIRNCFHCYCKSCF 644
+ ++G C C+ + C+
Sbjct: 441 VVQKKDGCDWIRCTVCHTEICW 462
>sp|Q9BYM8|UB7I3_HUMAN Ubiquitin-conjugating enzyme 7-interacting protein 3 (Hepatitis B virus X-associated protein 4) (HBV-associated factor 4) (RING finger protein 54) Length = 500 Score = 105 bits (261), Expect = 4e-22 Identities = 81/310 (26%), Positives = 136/310 (43%), Gaps = 46/310 (14%) Frame = +3 Query: 369 CQFCDQPN------CLKCEKRHYPNQCGDLNHNEDAIEAAAKVAMSRHRREALEEQMKNN 530 C F ++P C + Y E+ A + R ++ ++Q + N Sbjct: 192 CTFINKPTRPGCEMCCRARPEAYQVPASYQPDEEERARLAGEEEALRQYQQRKQQQQEGN 251 Query: 531 ----------AYIPNSESFECGICNSFILpreGLIIRNCFHCYCKSCFEETLKNVNFTSN 680 + + N+E EC +C S + P E +++R C H +C+ C + T++N + + Sbjct: 252 YLQHVQLDQRSLVLNTEPAECPVCYSVLAPGEAVVLRECLHTFCRECLQGTIRN-SQEAE 310 Query: 681 FKCP------KCGIEINNQEIIKAVGVEKL-----IGMACKE-AAPKMFCCKYPDCDG-- 818 CP C ++ +EI + E +G++ E + + CK PDC G Sbjct: 311 VSCPFIDNTYSCSGKLLEREIKALLTPEDYQRFLDLGISIAENRSAFSYHCKTPDCKGWC 370 Query: 819 MWDNDVIRKGSFICPVCKHSNCLDCEDVHEDKSCEEFR-----LTERYIDAQKA----KK 971 +++DV F CPVC H NCL C+ +HE +C+E++ + + A++ K Sbjct: 371 FFEDDV---NEFTCPVCFHVNCLLCKAIHEQMNCKEYQEDLALRAQNDVAARQTTEMLKV 427 Query: 972 KLQGESGIKCICGRFLVQ-KGGCGALHCDSCGRQICVFTKKIA-GNNGP-DTCGCTPTKT 1142 LQ ++C + +VQ K GC + C C +IC TK G GP DT G + Sbjct: 428 MLQQGEAMRCPQCQIVVQKKDGCDWIRCTVCHTEICWVTKGPRWGPGGPGDTSGGCRCRV 487 Query: 1143 ----CHPDCK 1160 CHP C+ Sbjct: 488 NGIPCHPSCQ 497
Score = 64.7 bits (156), Expect = 6e-10
Identities = 47/201 (23%), Positives = 84/201 (41%), Gaps = 7/201 (3%)
Frame = +3
Query: 63 CMKCKGTGIVDDSFTLRLCNHMFCHKCLCEYIKFHINCKIKCP-VEN---CNKNISDQII 230
C C ++ LR C H FC +CL I+ ++ CP ++N C+ + ++ I
Sbjct: 272 CPVCYSVLAPGEAVVLRECLHTFCRECLQGTIRNSQEAEVSCPFIDNTYSCSGKLLEREI 331
Query: 231 RKCLSNPEIALYEQ--ISLCKSTKS-ARKCLTSNCSAIMFIEDNKAIFQCQFCDQPNCLK 401
+ L+ + + IS+ ++ + + C T +C F ED+ F C C NCL
Sbjct: 332 KALLTPEDYQRFLDLGISIAENRSAFSYHCKTPDCKGWCFFEDDVNEFTCPVCFHVNCLL 391
Query: 402 CEKRHYPNQCGDLNHNEDAIEAAAKVAMSRHRREALEEQMKNNAYIPNSESFECGICNSF 581
C+ H C + + ED A +R E L+ ++ E+ C C
Sbjct: 392 CKAIHEQMNCKE--YQEDLALRAQNDVAARQTTEMLKVMLQ------QGEAMRCPQCQIV 443
Query: 582 ILpreGLIIRNCFHCYCKSCF 644
+ ++G C C+ + C+
Sbjct: 444 VQKKDGCDWIRCTVCHTEICW 464
>sp|Q62921|UB7I3_RAT Ubiquitin-conjugating enzyme 7-interacting protein 3 (RBCC protein interacting with PKC) (Protein kinase C-binding protein beta-15) Length = 498 Score = 105 bits (261), Expect = 4e-22 Identities = 69/244 (28%), Positives = 114/244 (46%), Gaps = 29/244 (11%) Frame = +3 Query: 516 QMKNNAYIPNSESFECGICNSFILpreGLIIRNCFHCYCKSCFEETLKNVN--------F 671 Q++ + + N+E EC +C S + P E +++R C H +C+ C + T++N Sbjct: 255 QLEQRSLVLNTEPAECPVCYSVLAPGEAVVLRECLHTFCRECLQGTIRNSQEAEVSCPFI 314 Query: 672 TSNFKCPKCGIEINNQEIIKAVGVEKLIGMACKEAAPK---MFCCKYPDCDG--MWDNDV 836 + + CP +E + ++ ++ + + A + + CK PDC G +++DV Sbjct: 315 DNTYSCPGKLLEREIRALLSPEDYQRFLDLGVSIAENRSTLSYHCKTPDCRGWCFFEDDV 374 Query: 837 IRKGSFICPVCKHSNCLDCEDVHEDKSCEEFR--LTERY---IDAQKAKKKL-----QGE 986 F CPVC NCL C+ +HE +C E++ L R + AQ+ + L QGE Sbjct: 375 ---NEFTCPVCTRVNCLLCKAIHERMNCREYQDDLAHRARNDVAAQQTTEMLRVMLQQGE 431 Query: 987 SGIKCICGRFLVQKGGCGALHCDSCGRQICVFTKKIA-GNNGP-DTCGCTPTKT----CH 1148 + C + +K GC + C C +IC TK G GP DT G + CH Sbjct: 432 AMYCPQCRIVVQKKDGCDWIRCTVCHTEICWVTKGPRWGPGGPGDTSGGCRCRVNGIPCH 491 Query: 1149 PDCK 1160 P C+ Sbjct: 492 PSCQ 495
Score = 63.9 bits (154), Expect = 1e-09
Identities = 48/202 (23%), Positives = 82/202 (40%), Gaps = 8/202 (3%)
Frame = +3
Query: 63 CMKCKGTGIVDDSFTLRLCNHMFCHKCLCEYIKFHINCKIKCP-VEN---CNKNISDQII 230
C C ++ LR C H FC +CL I+ ++ CP ++N C + ++ I
Sbjct: 270 CPVCYSVLAPGEAVVLRECLHTFCRECLQGTIRNSQEAEVSCPFIDNTYSCPGKLLEREI 329
Query: 231 RKCLSNPEIALYEQISLC----KSTKSARKCLTSNCSAIMFIEDNKAIFQCQFCDQPNCL 398
R LS + + + + +ST S C T +C F ED+ F C C + NCL
Sbjct: 330 RALLSPEDYQRFLDLGVSIAENRSTLSYH-CKTPDCRGWCFFEDDVNEFTCPVCTRVNCL 388
Query: 399 KCEKRHYPNQCGDLNHNEDAIEAAAKVAMSRHRREALEEQMKNNAYIPNSESFECGICNS 578
C+ H C + + +D A ++ E L ++ E+ C C
Sbjct: 389 LCKAIHERMNCRE--YQDDLAHRARNDVAAQQTTEMLRVMLQ------QGEAMYCPQCRI 440
Query: 579 FILpreGLIIRNCFHCYCKSCF 644
+ ++G C C+ + C+
Sbjct: 441 VVQKKDGCDWIRCTVCHTEICW 462
>sp|Q924T7|RNF31_MOUSE RING finger protein 31 Length = 1066 Score = 58.5 bits (140), Expect = 4e-08 Identities = 62/245 (25%), Positives = 98/245 (40%), Gaps = 36/245 (14%) Frame = +3 Query: 522 KNNAYIPNSESFECGICNSFILpreGL-IIRNCFHCYCKSCFEE----TLKNVNFTSNFK 686 ++ A++ + EC +C + LPR + + +C C CF + LK + T + Sbjct: 680 QDRAFLRRLLAQECAVCG-WALPRNRMQALISCECTICPECFRQHFTIALKEKHIT-DMV 737 Query: 687 CPKCG----------------IEINNQEIIKAVGV---EKLIGMACKEAAPKMFCCKYPD 809 CP CG ++I +E + K + A PK C Sbjct: 738 CPACGRPDLTDDAQLLSYFSTLDIQLRESLDPDAYALFHKKLTEAVLMRDPKFLWCAQCS 797 Query: 810 CDGMWDNDVIRKGSFICPVCKHSNCLDC----EDVHEDKSCEEFRLTERYID----AQKA 965 +++ + + CP C + C+ C E+ H +SCE+F+ +R D AQ Sbjct: 798 FGFIYEREQLEA---TCPQCHQTFCVRCKRQWEEQHRGRSCEDFQNWKRTNDPEYQAQGL 854 Query: 966 KKKLQGESGIKCICGRF--LVQKGGCGALHCDSCGRQIC--VFTKKIAGNNGPDTCGCTP 1133 LQ E+GI C +F + +GGC HC C Q C + A N PD C Sbjct: 855 AMYLQ-ENGIDCPKCKFSYALARGGCMHFHCTQCRHQFCSGCYNAFYAKNKCPDP-NCKV 912 Query: 1134 TKTCH 1148 K+ H Sbjct: 913 KKSLH 917
Score = 32.0 bits (71), Expect = 4.4
Identities = 30/133 (22%), Positives = 46/133 (34%), Gaps = 8/133 (6%)
Frame = +3
Query: 318 SNCSAIMFIEDNKAIFQCQFCDQPNCLKC----EKRHYPNQCGDLNHNEDAIEAAAKVAM 485
+ CS E + C C Q C++C E++H C D + +
Sbjct: 794 AQCSFGFIYEREQLEATCPQCHQTFCVRCKRQWEEQHRGRSCEDFQNWK----------- 842
Query: 486 SRHRREALEEQMKNNAYIPNSESFECGICN-SFILpreGLIIRNCFHC---YCKSCFEET 653
R E Q + A +C C S+ L R G + +C C +C C+
Sbjct: 843 ---RTNDPEYQAQGLAMYLQENGIDCPKCKFSYALARGGCMHFHCTQCRHQFCSGCYNA- 898
Query: 654 LKNVNFTSNFKCP 692
F + KCP
Sbjct: 899 -----FYAKNKCP 906
>sp|Q96EP0|RNF31_HUMAN RING finger protein 31 (Zinc in-between-RING-finger ubiquitin-associated domain protein) Length = 1072 Score = 56.6 bits (135), Expect = 2e-07 Identities = 60/246 (24%), Positives = 101/246 (41%), Gaps = 37/246 (15%) Frame = +3 Query: 522 KNNAYIPNSESFECGICNSFILpreGLIIRNCFHCYCKSCFEE----TLKNVNFTSNFKC 689 ++ A++ + EC +C + + +C C CF + LK + T + C Sbjct: 686 QDRAFLRRLLAQECAVCGWALPHNRMQALTSCECTICPDCFRQHFTIALKEKHIT-DMVC 744 Query: 690 PKCG----------------IEINNQEIIK----AVGVEKLI-GMACKEAAPKMFCCKYP 806 P CG ++I +E ++ A+ +KL G+ ++ PK C Sbjct: 745 PACGRPDLTDDTQLLSYFSTLDIQLRESLEPDAYALFHKKLTEGVLMRD--PKFLWCAQC 802 Query: 807 DCDGMWDNDVIRKGSFICPVCKHSNCLDC----EDVHEDKSCEEFRLTERYID----AQK 962 +++ + + CP C + C+ C E+ H +SCE+F+ +R D AQ Sbjct: 803 SFGFIYEREQLEA---TCPQCHQTFCVRCKRQWEEQHRGRSCEDFQNWKRMNDPEYQAQG 859 Query: 963 AKKKLQGESGIKCICGRF--LVQKGGCGALHCDSCGRQIC--VFTKKIAGNNGPDTCGCT 1130 LQ E+GI C +F + +GGC HC C Q C + A N P+ C Sbjct: 860 LAMYLQ-ENGIDCPKCKFSYALARGGCMHFHCTQCRHQFCSGCYNAFYAKNKCPEP-NCR 917 Query: 1131 PTKTCH 1148 K+ H Sbjct: 918 VKKSLH 923
Score = 32.3 bits (72), Expect = 3.4
Identities = 30/134 (22%), Positives = 47/134 (35%), Gaps = 8/134 (5%)
Frame = +3
Query: 318 SNCSAIMFIEDNKAIFQCQFCDQPNCLKC----EKRHYPNQCGDLNHNEDAIEAAAKVAM 485
+ CS E + C C Q C++C E++H C D + +
Sbjct: 800 AQCSFGFIYEREQLEATCPQCHQTFCVRCKRQWEEQHRGRSCEDFQNWK----------- 848
Query: 486 SRHRREALEEQMKNNAYIPNSESFECGICN-SFILpreGLIIRNCFHC---YCKSCFEET 653
R E Q + A +C C S+ L R G + +C C +C C+
Sbjct: 849 ---RMNDPEYQAQGLAMYLQENGIDCPKCKFSYALARGGCMHFHCTQCRHQFCSGCYNA- 904
Query: 654 LKNVNFTSNFKCPK 695
F + KCP+
Sbjct: 905 -----FYAKNKCPE 913
>sp|Q8IWT3|PARC_HUMAN p53-associated parkin-like cytoplasmic protein (UbcH7 associated protein 1) Length = 2517 Score = 55.1 bits (131), Expect = 5e-07 Identities = 54/229 (23%), Positives = 83/229 (36%), Gaps = 19/229 (8%) Frame = +3 Query: 33 AQFIVI*MSICMKCKGTGIVDDSFTLRLCNHMFCHKCLCEYIKFHI------NCKIKCPV 194 AQ + + C C DD C H C C EY+ I NC CP+ Sbjct: 2060 AQAVPVRPDHCPVCVSPLGCDDDLPSLCCMHYCCKSCWNEYLTTRIEQNLVLNC--TCPI 2117 Query: 195 ENCNKNISDQIIRKCLSNPE-IALYEQISL---CKSTKSARKCLT-SNCSAIMFIEDNKA 359 +C + IR +S+PE I+ YE+ L +S + C C I+ + Sbjct: 2118 ADCPAQPTGAFIRAIVSSPEVISKYEKALLRGYVESCSNLTWCTNPQGCDRILCRQGLGC 2177 Query: 360 IFQCQFCDQPNCLKCE--KRHYPNQCGDLNHNED---AIEAAAKVAMSRHRREALEEQMK 524 C C +C C + HYP CG ++ D + + A S+H + + ++ Sbjct: 2178 GTTCSKCGWASCFNCSFPEAHYPASCGHMSQWVDDGGYYDGMSVEAQSKHLAKLISKR-- 2235 Query: 525 NNAYIPNSESFECGICNSFILpreGLIIRNCFHC---YCKSCFEETLKN 662 C C + I EG + C C +C C + N Sbjct: 2236 ------------CPSCQAPIEKNEGCLHMTCAKCNHGFCWRCLKSWKPN 2272
>sp|Q80TT8|PARC_MOUSE p53-associated parkin-like cytoplasmic protein Length = 1865 Score = 54.7 bits (130), Expect = 6e-07 Identities = 51/204 (25%), Positives = 81/204 (39%), Gaps = 19/204 (9%) Frame = +3 Query: 93 DDSFTLRLCNHMFCHKCLCEYI------KFHINCKIKCPVENCNKNISDQIIRKCLSNPE 254 DDS +L C H C C EY+ F +NC CP+ +C + IR +S+PE Sbjct: 1420 DDSPSL-CCLHCCCKSCWNEYLTTRIEQNFVLNCT--CPIADCPAQPTGAFIRNIVSSPE 1476 Query: 255 -IALYEQISL---CKSTKSARKCLT-SNCSAIMFIEDNKAIFQCQFCDQPNCLKCE--KR 413 I+ YE+ L +S + C C I+ + + C C +C C + Sbjct: 1477 VISKYEKALLRGYVESCSNLTWCTNPQGCDRILCRQGLGSGTTCSKCGWASCFSCSFPEA 1536 Query: 414 HYPNQCGDLNHNED---AIEAAAKVAMSRHRREALEEQMKNNAYIPNSESFECGICNSFI 584 HYP CG ++ D + + A S+H + + ++ C C + I Sbjct: 1537 HYPASCGHMSQWVDDGGYYDGMSVEAQSKHLAKLISKR--------------CPSCQAPI 1582 Query: 585 LpreGLIIRNCFHC---YCKSCFE 647 EG + C C +C C + Sbjct: 1583 EKNEGCLHMTCARCNHGFCWRCLK 1606
Score = 43.1 bits (100), Expect = 0.002
Identities = 45/200 (22%), Positives = 73/200 (36%), Gaps = 27/200 (13%)
Frame = +3
Query: 558 ECGICNSFILpreGLIIRNCFHCYCKSCFEETLK---NVNFTSNFKCP--KCGIE----- 707
+C +C + + P + C HC CKSC+ E L NF N CP C +
Sbjct: 1408 QCPVCVTPLGPHDDSPSLCCLHCCCKSCWNEYLTTRIEQNFVLNCTCPIADCPAQPTGAF 1467
Query: 708 ----INNQEIIKAVGVEKLIGMACKEAAPKMFCCKYPD-CDGMWDNDVIRKGSFICPVCK 872
+++ E+I EK + E+ + C P CD + + G+ C C
Sbjct: 1468 IRNIVSSPEVISK--YEKALLRGYVESCSNLTWCTNPQGCDRILCRQGLGSGT-TCSKCG 1524
Query: 873 HSNCLDCE--DVHEDKSCEEFRLTERYID---------AQKAKKKLQGESGIKC-ICGRF 1016
++C C + H SC +++D + K L +C C
Sbjct: 1525 WASCFSCSFPEAHYPASCGHM---SQWVDDGGYYDGMSVEAQSKHLAKLISKRCPSCQAP 1581
Query: 1017 LVQKGGCGALHCDSCGRQIC 1076
+ + GC + C C C
Sbjct: 1582 IEKNEGCLHMTCARCNHGFC 1601
>sp|Q925F3|UB7I4_MOUSE Ubiquitin-conjugating enzyme 7-interacting protein 4 (UbcM4-interacting protein 4) (RING finger protein 144) Length = 292 Score = 51.6 bits (122), Expect = 5e-06 Identities = 49/212 (23%), Positives = 80/212 (37%), Gaps = 17/212 (8%) Frame = +3 Query: 63 CMKCKGTGIVDDSFTLRLCNHMFCHKCLCEY----IKFHINCKIKCPVENCNK--NISDQ 224 C C G + T+ C +FC CL +Y IK + I CP C K ++ + Sbjct: 20 CKLCLGEYPAEQMTTIAQCQCIFCTLCLKQYVELLIKEGLETAISCPDAACPKQGHLQEN 79 Query: 225 IIRKCLSNPEIALYEQISLCKST---KSARKCLTSNCSAIMFIED----NKAIFQCQFCD 383 I ++ + Y+++ + C S C A+ ++D + QC+ CD Sbjct: 80 EIECMVAAEIMQRYKKLQFEREVLFDPCRTWCPASTCQAVCQLQDIGLQTPQLVQCKACD 139 Query: 384 QPNCLKCEKRHYPNQ-CGDLNHNEDAIEAAAKVAMSRHRREALEEQMKNNAYIPNSESFE 560 C C+ R +P Q C + I S + E + +K Sbjct: 140 MEFCSACKARWHPGQGCPETM----PITFLPGETSSAFKMEEGDAPIK-----------R 184 Query: 561 CGICNSFILpreG---LIIRNCFHCYCKSCFE 647 C C +I EG ++ +NC H +C C E Sbjct: 185 CPKCRVYIERDEGCAQMMCKNCKHAFCWYCLE 216
>sp|P58283|UB7I1_MOUSE E3 ubiquitin ligase TRIAD3 (Ubiquitin-conjugating enzyme 7-interacting protein 1) (UbcM4-interacting protein 83) Length = 853 Score = 50.1 bits (118), Expect = 2e-05 Identities = 53/220 (24%), Positives = 80/220 (36%), Gaps = 22/220 (10%) Frame = +3 Query: 483 MSRHRREALEEQMKNNAYIPNSESFECGICNSFILpreGLIIRNCFHCYCKSCFEETLKN 662 M+ H L QM Y + + EC C P E L H +CK C + Sbjct: 477 MAEHEDFLLALQMNEEQYQKDGQLIECRCCYGEF-PFEELTQCADAHLFCKECLIRYAQE 535 Query: 663 VNF---TSNFKCPK--CGIEINNQEIIKAVG-----------VEKLIGMACKEAAPKMFC 794 F S C + C E+ K + E+ + A + + Sbjct: 536 AVFGSGKSELSCMEGSCTCSFPTSELEKVLPQTILYKYYERKAEEEVAAAYADELVRCPS 595 Query: 795 CKYPDCDGMWDNDVIRKGSFICPV--CKHSNCLDCEDV---HEDKSCEEFRLTERYIDAQ 959 C +P + D+DV R F CP C+ C C+ + H +CEE + Sbjct: 596 CSFP---ALLDSDVKR---FSCPNPRCRKETCRKCQGLWKEHNGLTCEELAEKDDIKYRT 649 Query: 960 KAKKKLQGESGIKCI-CGRFLVQKGGCGALHCDSCGRQIC 1076 ++K+ KC CG L++ GC + C CG Q+C Sbjct: 650 SIEEKMTAARIRKCHKCGTGLIKSEGCNRMSC-RCGAQMC 688
Score = 34.7 bits (78), Expect = 0.68
Identities = 39/191 (20%), Positives = 73/191 (38%), Gaps = 17/191 (8%)
Frame = +3
Query: 123 HMFCHKCLCEYIKFHI----NCKIKCPVENCNKNISDQIIRKCLSNPEIALY-------- 266
H+FC +CL Y + + ++ C +C + + K L P+ LY
Sbjct: 522 HLFCKECLIRYAQEAVFGSGKSELSCMEGSCTCSFPTSELEKVL--PQTILYKYYERKAE 579
Query: 267 EQISLCKSTKSARKCLTSNCSAIMFIEDNKAIFQC--QFCDQPNCLKCE---KRHYPNQC 431
E+++ + + R +CS ++ + F C C + C KC+ K H C
Sbjct: 580 EEVAAAYADELVR---CPSCSFPALLDSDVKRFSCPNPRCRKETCRKCQGLWKEHNGLTC 636
Query: 432 GDLNHNEDAIEAAAKVAMSRHRREALEEQMKNNAYIPNSESFECGICNSFILpreGLIIR 611
+L +D R ++EE+M + +C C + ++ EG
Sbjct: 637 EELAEKDDI-----------KYRTSIEEKM------TAARIRKCHKCGTGLIKSEGCNRM 679
Query: 612 NCFHCYCKSCF 644
+C C + C+
Sbjct: 680 SC-RCGAQMCY 689
>sp|Q9NWF9|UB7I1_HUMAN E3 ubiquitin ligase TRIAD3 (Ubiquitin conjugating enzyme 7 interacting protein 1) (Zinc finger protein inhibiting NF-kappa-B) Length = 866 Score = 49.7 bits (117), Expect = 2e-05 Identities = 53/220 (24%), Positives = 80/220 (36%), Gaps = 22/220 (10%) Frame = +3 Query: 483 MSRHRREALEEQMKNNAYIPNSESFECGICNSFILpreGLIIRNCFHCYCKSCFEETLKN 662 M+ H L QM Y + + EC C P E L H +CK C + Sbjct: 489 MAEHEDFLLALQMNEEQYQKDGQLIECRCCYGEF-PFEELTQCADAHLFCKECLIRYAQE 547 Query: 663 VNFTSN---FKCPK--CGIEINNQEIIKAVG-----------VEKLIGMACKEAAPKMFC 794 F S C + C E+ K + E+ + A + + Sbjct: 548 AVFGSGKLELSCMEGSCTCSFPTSELEKVLPQTILYKYYERKAEEEVAAAYADELVRCPS 607 Query: 795 CKYPDCDGMWDNDVIRKGSFICPV--CKHSNCLDCEDV---HEDKSCEEFRLTERYIDAQ 959 C +P + D+DV R F CP C+ C C+ + H +CEE + Sbjct: 608 CSFP---ALLDSDVKR---FSCPNPHCRKETCRKCQGLWKEHNGLTCEELAEKDDIKYRT 661 Query: 960 KAKKKLQGESGIKCI-CGRFLVQKGGCGALHCDSCGRQIC 1076 ++K+ KC CG L++ GC + C CG Q+C Sbjct: 662 SIEEKMTAARIRKCHKCGTGLIKSEGCNRMSC-RCGAQMC 700
Score = 35.4 bits (80), Expect = 0.40
Identities = 39/191 (20%), Positives = 73/191 (38%), Gaps = 17/191 (8%)
Frame = +3
Query: 123 HMFCHKCLCEYIKFHI----NCKIKCPVENCNKNISDQIIRKCLSNPEIALY-------- 266
H+FC +CL Y + + ++ C +C + + K L P+ LY
Sbjct: 534 HLFCKECLIRYAQEAVFGSGKLELSCMEGSCTCSFPTSELEKVL--PQTILYKYYERKAE 591
Query: 267 EQISLCKSTKSARKCLTSNCSAIMFIEDNKAIFQC--QFCDQPNCLKCE---KRHYPNQC 431
E+++ + + R +CS ++ + F C C + C KC+ K H C
Sbjct: 592 EEVAAAYADELVR---CPSCSFPALLDSDVKRFSCPNPHCRKETCRKCQGLWKEHNGLTC 648
Query: 432 GDLNHNEDAIEAAAKVAMSRHRREALEEQMKNNAYIPNSESFECGICNSFILpreGLIIR 611
+L +D R ++EE+M + +C C + ++ EG
Sbjct: 649 EELAEKDDI-----------KYRTSIEEKM------TAARIRKCHKCGTGLIKSEGCNRM 691
Query: 612 NCFHCYCKSCF 644
+C C + C+
Sbjct: 692 SC-RCGAQMCY 701
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 149,821,659
Number of Sequences: 369166
Number of extensions: 3133473
Number of successful extensions: 9360
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 8291
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 9282
length of database: 68,354,980
effective HSP length: 114
effective length of database: 47,295,190
effective search space used: 16553316500
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail