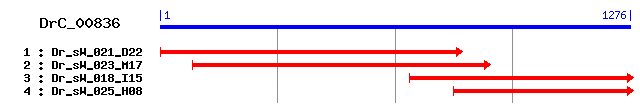
DrC_00836
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00836 (1276 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P40529|AGE2_YEAST Protein AGE2 119 2e-26 sp|Q96P47|CENG3_HUMAN Centaurin-gamma 3 106 2e-22 sp|Q96P50|CENB5_HUMAN Centaurin-beta 5 (Cnt-b5) 104 5e-22 sp|Q8BXK8|CENG2_MOUSE Centaurin-gamma 2 (ARF-GAP with GTP-b... 104 5e-22 sp|Q9UPQ3|CENG2_HUMAN Centaurin-gamma 2 (ARF-GAP with GTP-b... 103 1e-21 sp|Q15057|CENB2_HUMAN Centaurin-beta 2 (Cnt-b2) 102 2e-21 sp|Q9NGC3|CEG1A_DROME Centaurin-gamma 1A 102 3e-21 sp|Q15027|CENB1_HUMAN Centaurin-beta 1 (Cnt-b1) 100 9e-21 sp|O75689|CENA1_HUMAN Centaurin-alpha 1 (Putative MAPK-acti... 99 3e-20 sp|O74345|UCP3_SCHPO UBA-domain containing protein 3 98 4e-20
>sp|P40529|AGE2_YEAST Protein AGE2 Length = 298 Score = 119 bits (298), Expect = 2e-26 Identities = 99/357 (27%), Positives = 163/357 (45%), Gaps = 9/357 (2%) Frame = +3 Query: 27 LAELLKDDDNKYCVDCDSK-GPRWASWTLGIFLCIRCAGIHRNLGVHLSRVKSVNLDSWT 203 L+ LL+D N +C DC ++ PRWASW+LG+F+CI+CAGIHR+LG H+S+VKSV+LD+W Sbjct: 11 LSALLRDPGNSHCADCKAQLHPRWASWSLGVFICIKCAGIHRSLGTHISKVKSVDLDTWK 70 Query: 204 PQQVAMMKEIGNS-RGRAIYEANLEDNFR-RPQTD-SSLEQFIRSKYEHKRYIAKEWIPP 374 + + + + N+ R + YEA L D + R TD SSL+ FI++KYE+K+ WI Sbjct: 71 EEHLVKLIQFKNNLRANSYYEATLADELKQRKITDTSSLQNFIKNKYEYKK-----WIGD 125 Query: 375 TPTVDVIAEEIRKLEQNQNKKRVNYNNTVTNAIIPLS----RRDNIETPGNIKSIEIDDK 542 +++ + + E +K N++ +NA + S ++ + P ++ S + Sbjct: 126 LSSIEGLND---STEPVLHKPSANHSLPASNARLDQSSNSLQKTQTQPPSHLLSTSRSNT 182 Query: 543 SQPVVNNKIDLLGLESDNVTNNIDDAFGDFVFAAQPTISTCQTVTGSATKPVTVSSAVDD 722 S ++N ++ L + N +VT SAT ++ +A Sbjct: 183 S--LLNLQVSSLSKTTSNT-----------------------SVTSSAT---SIGAA--- 211 Query: 723 XXXXXXXXXXXXXXXPGNNIAPSVSTSSAIPNISGKLD-KSSILALYNNSSSTMNTMGQM 899 + T + + + D K SIL+LY+ S+ + Sbjct: 212 ----------------------NTKTGNRVGEFGQRNDLKKSILSLYSKPSAQTQSQNSF 249 Query: 900 PNSTTWPNAGAPAYQSNLMQSDFQSQGYAMSGNNWMVPGQMNANSAFGVQHNPFFNN 1070 STT P S F + G + NN M N++S + N F N Sbjct: 250 FTSTTPQPCNTP--------SPFVNTGITATNNNSM---NSNSSSNISLDDNELFKN 295
>sp|Q96P47|CENG3_HUMAN Centaurin-gamma 3 Length = 876 Score = 106 bits (264), Expect = 2e-22 Identities = 50/116 (43%), Positives = 76/116 (65%), Gaps = 3/116 (2%) Frame = +3 Query: 54 NKYCVDCDSKGPRWASWTLGIFLCIRCAGIHRNLGVHLSRVKSVNLDSWTPQQVAMMKEI 233 N +C+DCD+ P WAS LG +CI C+GIHR+LG HLSRV+S++LD W P+ +A+M + Sbjct: 639 NSFCIDCDAPNPDWASLNLGALMCIECSGIHRHLGAHLSRVRSLDLDDWPPELLAVMTAM 698 Query: 234 GNSRGRAIYEANLEDNFRRPQTDS---SLEQFIRSKYEHKRYIAKEWIPPTPTVDV 392 GN+ +++E L + +P D+ E++IR+KYE K ++A P P+ DV Sbjct: 699 GNALANSVWEGAL-GGYSKPGPDACREEKERWIRAKYEQKLFLA-----PLPSSDV 748
>sp|Q96P50|CENB5_HUMAN Centaurin-beta 5 (Cnt-b5) Length = 759 Score = 104 bits (260), Expect = 5e-22 Identities = 52/111 (46%), Positives = 68/111 (61%), Gaps = 4/111 (3%) Frame = +3 Query: 54 NKYCVDCDSKGPRWASWTLGIFLCIRCAGIHRNLGVHLSRVKSVNLDSWTPQQVAMMKEI 233 N C DC PRWAS LG+ LCI C+GIHR+LGVH S+V+S+ LDSW P+ + +M E+ Sbjct: 373 NSQCGDCGQPDPRWASINLGVLLCIECSGIHRSLGVHCSKVRSLTLDSWEPELLKLMCEL 432 Query: 234 GNSRGRAIYEANLEDNFRRPQTDSSLEQ----FIRSKYEHKRYIAKEWIPP 374 GNS IYEA E R T SS Q +I+ KY K+++ K + P Sbjct: 433 GNSAVNQIYEAQCEGAGSRKPTASSSRQDKEAWIKDKYVEKKFLRKAPMAP 483
>sp|Q8BXK8|CENG2_MOUSE Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP1) Length = 857 Score = 104 bits (260), Expect = 5e-22 Identities = 47/104 (45%), Positives = 69/104 (66%), Gaps = 3/104 (2%) Frame = +3 Query: 54 NKYCVDCDSKGPRWASWTLGIFLCIRCAGIHRNLGVHLSRVKSVNLDSWTPQQVAMMKEI 233 N +CVDCD++ P WAS LG +CI C+GIHRNLG HLSRV+S++LD W + + +M I Sbjct: 621 NSHCVDCDTQNPNWASLNLGALMCIECSGIHRNLGTHLSRVRSLDLDDWPMELIKVMSSI 680 Query: 234 GNSRGRAIYEANLEDNFRRPQTDSSLEQ---FIRSKYEHKRYIA 356 GN +++E + +P DS+ E+ +IR+KYE K ++A Sbjct: 681 GNELANSVWEEGSQGR-TKPSLDSTREEKERWIRAKYEQKLFLA 723
>sp|Q9UPQ3|CENG2_HUMAN Centaurin-gamma 2 (ARF-GAP with GTP-binding protein-like, ankyrin repeat and pleckstrin homology domains 1) (AGAP1) Length = 857 Score = 103 bits (257), Expect = 1e-21 Identities = 46/104 (44%), Positives = 70/104 (67%), Gaps = 3/104 (2%) Frame = +3 Query: 54 NKYCVDCDSKGPRWASWTLGIFLCIRCAGIHRNLGVHLSRVKSVNLDSWTPQQVAMMKEI 233 N +CVDC+++ P WAS LG +CI C+GIHRNLG HLSRV+S++LD W + + +M I Sbjct: 621 NSHCVDCETQNPNWASLNLGALMCIECSGIHRNLGTHLSRVRSLDLDDWPIELIKVMSSI 680 Query: 234 GNSRGRAIYEANLEDNFRRPQTDSSLEQ---FIRSKYEHKRYIA 356 GN +++E + + +P DS+ E+ +IR+KYE K ++A Sbjct: 681 GNELANSVWEESSQGR-TKPSVDSTREEKERWIRAKYEQKLFLA 723
>sp|Q15057|CENB2_HUMAN Centaurin-beta 2 (Cnt-b2) Length = 778 Score = 102 bits (255), Expect = 2e-21 Identities = 50/112 (44%), Positives = 72/112 (64%), Gaps = 3/112 (2%) Frame = +3 Query: 54 NKYCVDCDSKGPRWASWTLGIFLCIRCAGIHRNLGVHLSRVKSVNLDSWTPQQVAMMKEI 233 N C DC PRWAS LGI LCI C+GIHR+LGVH S+V+S+ LD+W P+ + +M E+ Sbjct: 411 NASCCDCGLADPRWASINLGITLCIECSGIHRSLGVHFSKVRSLTLDTWEPELLKLMCEL 470 Query: 234 GNSRGRAIYEANLED-NFRRPQTDSSLEQ--FIRSKYEHKRYIAKEWIPPTP 380 GN +YEAN+E ++PQ E+ +IR+KY ++++ K I +P Sbjct: 471 GNDVINRVYEANVEKMGIKKPQPGQRQEKEAYIRAKYVERKFVDKYSISLSP 522
>sp|Q9NGC3|CEG1A_DROME Centaurin-gamma 1A Length = 995 Score = 102 bits (253), Expect = 3e-21 Identities = 61/174 (35%), Positives = 96/174 (55%), Gaps = 9/174 (5%) Frame = +3 Query: 15 LQIILAELLKDDDNKYCVDCDSKGPRWASWTLGIFLCIRCAGIHRNLGVHLSRVKSVNLD 194 L +LA + N +CVDC + P WAS LG+ +CI C+G+HRNLG H+S+V+S+ LD Sbjct: 701 LAAMLAIRQRVPGNGFCVDCGAPNPEWASLNLGVLMCIECSGVHRNLGSHISKVRSLGLD 760 Query: 195 SWTPQQVAMMKEIGNSRGRAIYEANLEDNFRRPQTDSS---LEQFIRSKYEHKRYI---- 353 W +++M IGNS +++E+N +P + +S E+++RSKYE K ++ Sbjct: 761 DWPSPHLSVMLAIGNSLANSVWESNTRQRV-KPTSQASREDKERWVRSKYEAKEFLTPLG 819 Query: 354 --AKEWIPPTPTVDVIAEEIRKLEQNQNKKRVNYNNTVTNAIIPLSRRDNIETP 509 + P+P +I IR ++ N + VTNA + S RD + TP Sbjct: 820 NGSSAHPSPSPGQQLIEAVIRADIKSIVSILANCPSEVTNANV--SARD-VRTP 870
>sp|Q15027|CENB1_HUMAN Centaurin-beta 1 (Cnt-b1) Length = 740 Score = 100 bits (249), Expect = 9e-21 Identities = 48/122 (39%), Positives = 75/122 (61%), Gaps = 1/122 (0%) Frame = +3 Query: 24 ILAELLKDDDNKYCVDCDSKGPRWASWTLGIFLCIRCAGIHRNLGVHLSRVKSVNLDSWT 203 ++A++ D N C DC P WAS LG+ LCI+C+GIHR+LGVH S+V+S+ LDSW Sbjct: 407 VVAQVQSVDGNAQCCDCREPAPEWASINLGVTLCIQCSGIHRSLGVHFSKVRSLTLDSWE 466 Query: 204 PQQVAMMKEIGNSRGRAIYEANLED-NFRRPQTDSSLEQFIRSKYEHKRYIAKEWIPPTP 380 P+ V +M E+GN IYEA +E ++P S ++ + + H +Y+ K+++ P Sbjct: 467 PELVKLMCELGNVIINQIYEARVEAMAVKKPGPSCSRQE--KEAWIHAKYVEKKFLTKLP 524 Query: 381 TV 386 + Sbjct: 525 EI 526
>sp|O75689|CENA1_HUMAN Centaurin-alpha 1 (Putative MAPK-activating protein PM25) Length = 374 Score = 98.6 bits (244), Expect = 3e-20 Identities = 51/113 (45%), Positives = 71/113 (62%), Gaps = 3/113 (2%) Frame = +3 Query: 33 ELLKDDDNKYCVDCDSKGPRWASWTLGIFLCIRCAGIHRNLGVHLSRVKSVNLDSWTPQQ 212 ELL+ N C DC + P WAS+TLG+F+C+ C+GIHRN+ +S+VKSV LD+W Q Sbjct: 11 ELLQRPGNARCADCGAPDPDWASYTLGVFICLSCSGIHRNI-PQVSKVKSVRLDAWEEAQ 69 Query: 213 VAMMKEIGNSRGRAIYEANLEDNFRRP-QTDSSL--EQFIRSKYEHKRYIAKE 362 V M GN RA +E+ + + RP +D L EQ+IR+KYE + +I E Sbjct: 70 VEFMASHGNDAARARFESKVPSFYYRPTPSDCQLLREQWIRAKYERQEFIYPE 122
>sp|O74345|UCP3_SCHPO UBA-domain containing protein 3 Length = 601 Score = 98.2 bits (243), Expect = 4e-20 Identities = 44/103 (42%), Positives = 62/103 (60%), Gaps = 3/103 (2%) Frame = +3 Query: 54 NKYCVDCDSKGPRWASWTLGIFLCIRCAGIHRNLGVHLSRVKSVNLDSWTPQQVAMMKEI 233 N C DC ++G +WASW LGIFLC+RCA IHR LG H+S+VKS++LD W+ Q+ MK Sbjct: 20 NNLCADCSTRGVQWASWNLGIFLCLRCATIHRKLGTHVSKVKSISLDEWSNDQIEKMKHW 79 Query: 234 GNSRGRAIYEANLEDN---FRRPQTDSSLEQFIRSKYEHKRYI 353 GN + N + + +E++IR KYE K ++ Sbjct: 80 GNINANRYWNPNPLSHPLPTNALSDEHVMEKYIRDKYERKLFL 122
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 139,760,309
Number of Sequences: 369166
Number of extensions: 2911390
Number of successful extensions: 7904
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 7392
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7869
length of database: 68,354,980
effective HSP length: 113
effective length of database: 47,479,925
effective search space used: 14766256675
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail