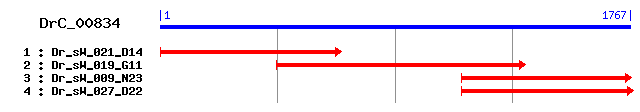
DrC_00834
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00834 (1767 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P42356|PI4KA_HUMAN Phosphatidylinositol 4-kinase alpha (... 372 e-102 sp|P37297|STT4_YEAST Phosphatidylinositol 4-kinase STT4 (PI... 275 2e-73 sp|Q9UBF8|PI4KB_HUMAN Phosphatidylinositol 4-kinase beta (P... 189 3e-47 sp|O08561|PI4KB_RAT Phosphatidylinositol 4-kinase beta (Ptd... 187 1e-46 sp|Q8BKC8|PI4KB_MOUSE Phosphatidylinositol 4-kinase beta (P... 187 1e-46 sp|P54677|PI4K_DICDI Phosphatidylinositol 4-kinase (PI4-kin... 186 2e-46 sp|O02810|PI4KB_BOVIN Phosphatidylinositol 4-kinase beta (P... 186 2e-46 sp|Q9UW24|PIK1A_CANAL Phosphatidylinositol 4-kinase PIK1a (... 158 4e-38 sp|Q9UW20|PIK1_CANAL Phosphatidylinositol 4-kinase PIK1alph... 158 4e-38 sp|P39104|PIK1_YEAST Phosphatidylinositol 4-kinase PIK1 (PI... 157 7e-38
>sp|P42356|PI4KA_HUMAN Phosphatidylinositol 4-kinase alpha (PI4-kinase) (PtdIns-4-kinase) (PI4K-alpha) Length = 2044 Score = 372 bits (954), Expect = e-102 Identities = 170/261 (65%), Positives = 221/261 (84%) Frame = +3 Query: 801 IYYQACIFKVGDDVRQDILALQVIRLFKNILEREGLDHYLYPYRVVATSPGCGVIECVPD 980 I +QA IFKVGDD RQD+LALQ+I LFKNI + GLD +++PYRVVAT+PGCGVIEC+PD Sbjct: 1784 ISWQAAIFKVGDDCRQDMLALQIIDLFKNIFQLVGLDLFVFPYRVVATAPGCGVIECIPD 1843 Query: 981 SKSRDQIGRTTDSGMYDYFLSLFGEETTPSYQEARRNFVMSMASYSVVCYLLQIKDRHNG 1160 SRDQ+GR TD GMYDYF +G+E+T ++Q+AR NF+ SMA+YS++ +LLQIKDRHNG Sbjct: 1844 CTSRDQLGRQTDFGMYDYFTRQYGDESTLAFQQARYNFIRSMAAYSLLLFLLQIKDRHNG 1903 Query: 1161 NIMIDKHGHIIHIDFGFMFESSPGNNMGFEPDIKLTKEMMMVMGGKLDSPAFQWFEQLSV 1340 NIM+DK GHIIHIDFGFMFESSPG N+G+EPDIKLT EM+M+MGGK+++ F+WF ++ V Sbjct: 1904 NIMLDKKGHIIHIDFGFMFESSPGGNLGWEPDIKLTDEMVMIMGGKMEATPFKWFMEMCV 1963 Query: 1341 QAYLVLRPYQEALVALVSLMLDSGLPCFRGQTIKLLRQRFTPLMSDRDAANHYLRVLRTC 1520 + YL +RPY +A+V+LV+LMLD+GLPCFRGQTIKLL+ RF+P M++R+AAN ++V+++C Sbjct: 1964 RGYLAVRPYMDAVVSLVTLMLDTGLPCFRGQTIKLLKHRFSPNMTEREAANFIMKVIQSC 2023 Query: 1521 CDHWRANTYDFIQYFQNQIPY 1583 R+ TYD IQY+QN IPY Sbjct: 2024 FLSNRSRTYDMIQYYQNDIPY 2044
Score = 295 bits (756), Expect = 2e-79
Identities = 139/224 (62%), Positives = 173/224 (77%), Gaps = 3/224 (1%)
Frame = +2
Query: 20 DIPELCYVPTWXXXXXXXXLSFFSRMYPQHPLTHQYAVKVLSLYPPDTLIFYVPQLVQGI 199
D PEL +V W LS+FS MYP HPLT QY VKVL +PPD ++FY+PQ+VQ +
Sbjct: 1544 DAPELSHVLCWAPTDPPTGLSYFSSMYPPHPLTAQYGVKVLRSFPPDAILFYIPQIVQAL 1603
Query: 200 RYDKMGYLTEFILEIVHLSPILAHQLIWNMKTNMYKDEDGQVKDPDIGLQLESVIDEINK 379
RYDKMGY+ E+IL S +LAHQ IWNMKTN+Y DE+G KDPDIG L+ +++EI
Sbjct: 1604 RYDKMGYVREYILWAASKSQLLAHQFIWNMKTNIYLDEEGHQKDPDIGDLLDQLVEEITG 1663
Query: 380 SFSGMSLSFSEREFDFFDKITNVSGVIRPYPKGQQRQQACLEALRKIELQPGCYLPSNPD 559
S SG + F +REFDFF+KITNVS +I+PYPKG +R++ACL AL ++++QPGCYLPSNP+
Sbjct: 1664 SLSGPAKDFYQREFDFFNKITNVSAIIKPYPKGDERKKACLSALSEVKVQPGCYLPSNPE 1723
Query: 560 AIVLEIDYLSGIPLQSAAKAPYLAKFKVKRCGVQKLEK---KCR 682
AIVL+IDY SG P+QSAAKAPYLAKFKVKRCGV +LEK +CR
Sbjct: 1724 AIVLDIDYKSGTPMQSAAKAPYLAKFKVKRCGVSELEKEGLRCR 1767
>sp|P37297|STT4_YEAST Phosphatidylinositol 4-kinase STT4 (PI4-kinase) (PtdIns-4-kinase) Length = 1900 Score = 275 bits (704), Expect = 2e-73 Identities = 145/282 (51%), Positives = 191/282 (67%), Gaps = 3/282 (1%) Frame = +3 Query: 747 THEIKENRKNNPID--NKDDIYYQACIFKVGDDVRQDILALQVIRLFKNILEREGLDHYL 920 T +IK++ K+ P+ NK+ +QA IFKVGDD RQD+LALQ+I LF+ I GLD Y+ Sbjct: 1620 TFKIKKDVKD-PLTGKNKEVEKWQAAIFKVGDDCRQDVLALQLISLFRTIWSSIGLDVYV 1678 Query: 921 YPYRVVATSPGCGVIECVPDSKSRDQIGRTTDSGMYDYFLSLFGEETTPSYQEARRNFVM 1100 +PYRV AT+PGCGVI+ +P+S SRD +GR +G+Y+YF S FG E+T +Q AR NFV Sbjct: 1679 FPYRVTATAPGCGVIDVLPNSVSRDMLGREAVNGLYEYFTSKFGNESTIEFQNARNNFVK 1738 Query: 1101 SMASYSVVCYLLQIKDRHNGNIMIDKHGHIIHIDFGFMFESSPGNNMGFEPDIKLTKEMM 1280 S+A YSV+ YLLQ KDRHNGNIM D GH +HIDFGF+F+ PG KLTKEM+ Sbjct: 1739 SLAGYSVISYLLQFKDRHNGNIMYDDQGHCLHIDFGFIFDIVPGGIKFEAVPFKLTKEMV 1798 Query: 1281 MVMGGKLDSPAFQWFEQLSVQAYLVLRPYQEALVALVSLMLDSGLPCFRG-QTIKLLRQR 1457 VMGG +PA+ FE+L ++AYL RP+ EA++ V+ ML SGLPCF+G +TI+ LR R Sbjct: 1799 KVMGGSPQTPAYLDFEELCIKAYLAARPHVEAIIECVNPMLGSGLPCFKGHKTIRNLRAR 1858 Query: 1458 FTPLMSDRDAANHYLRVLRTCCDHWRANTYDFIQYFQNQIPY 1583 F P +D +AA + ++R + YD Q N IPY Sbjct: 1859 FQPQKTDHEAALYMKALIRKSYESIFTKGYDEFQRLTNGIPY 1900
Score = 159 bits (402), Expect = 2e-38
Identities = 82/215 (38%), Positives = 129/215 (60%), Gaps = 2/215 (0%)
Frame = +2
Query: 50 WXXXXXXXXLSFFSRMYPQHPLTHQYAVKVLSLYPPDTLIFYVPQLVQGIRYDKMGYLTE 229
W ++ F + + Q+++ L + FYVPQ+VQ +RYDK GY+
Sbjct: 1425 WAPVSPLKSINLFLPEWQGNSFILQFSIYSLESQDVNLAFFYVPQIVQCLRYDKTGYVER 1484
Query: 230 FILEIVHLSPILAHQLIWNMKTNMYKDEDGQVKDPDIGLQLESVIDEINKSFSGMSLSFS 409
IL+ +S + +HQ+IWNM N YKD++G +++ +I L+ + + + SFS F
Sbjct: 1485 LILDTAKISVLFSHQIIWNMLANCYKDDEG-IQEDEIKPTLDRIRERMVSSFSQSHRDFY 1543
Query: 410 EREFDFFDKITNVSGVIRPYPKGQ--QRQQACLEALRKIELQPGCYLPSNPDAIVLEIDY 583
EREF+FFD++T +SG ++PY K +++ E + KIE++P YLPSNPD +V++ID
Sbjct: 1544 EREFEFFDEVTGISGKLKPYIKKSKAEKKHKIDEEMSKIEVKPDVYLPSNPDGVVIDIDR 1603
Query: 584 LSGIPLQSAAKAPYLAKFKVKRCGVQKLEKKCRDV 688
SG PLQS AKAP++A FK+K+ L K ++V
Sbjct: 1604 KSGKPLQSHAKAPFMATFKIKKDVKDPLTGKNKEV 1638
>sp|Q9UBF8|PI4KB_HUMAN Phosphatidylinositol 4-kinase beta (PtdIns 4-kinase) (PI4Kbeta) (PI4K-beta) (NPIK) (PI4K92) Length = 816 Score = 189 bits (479), Expect = 3e-47 Identities = 107/278 (38%), Positives = 164/278 (58%), Gaps = 3/278 (1%) Frame = +3 Query: 753 EIKENRKNNPIDNKDDIYYQACIFKVGDDVRQDILALQVIRLFKNILEREGLDHYLYPYR 932 +++ R+ +P + + + I K GDD+RQ++LA QV++ ++I E+E + ++ PY+ Sbjct: 540 KVRRIREGSPYGHLPNWRLLSVIVKCGDDLRQELLAFQVLKQLQSIWEQERVPLWIKPYK 599 Query: 933 VVATSPGCGVIECVPDSKSRDQIGRTTDSGMYDYFLSLFGEETTPSYQEARRNFVMSMAS 1112 ++ S G+IE V ++ S Q+ + + + DYFL G TT ++ A+RNFV S A Sbjct: 600 ILVISADSGMIEPVVNAVSIHQVKKQSQLSLLDYFLQEHGSYTTEAFLSAQRNFVQSCAG 659 Query: 1113 YSVVCYLLQIKDRHNGNIMIDKHGHIIHIDFGFMFESSPGNNMGFEPD-IKLTKEMMMVM 1289 Y +VCYLLQ+KDRHNGNI++D GHIIHIDFGF+ SSP N+GFE KLT E + VM Sbjct: 660 YCLVCYLLQVKDRHNGNILLDAEGHIIHIDFGFILSSSP-RNLGFETSAFKLTTEFVDVM 718 Query: 1290 GGKLDSPAFQWFEQLSVQAYLVLRPYQEALVALVSLMLD-SGLPCFRG-QTIKLLRQRFT 1463 GG LD F +++ L +Q + R + + +V +V +M S LPCF G TI+ L++RF Sbjct: 719 GG-LDGDMFNYYKMLMLQGLIAARKHMDKVVQIVEIMQQGSQLPCFHGSSTIRNLKERFH 777 Query: 1464 PLMSDRDAANHYLRVLRTCCDHWRANTYDFIQYFQNQI 1577 M++ +++ YD QY N I Sbjct: 778 MSMTEEQLQLLVEQMVDGSMRSITTKLYDGFQYLTNGI 815
>sp|O08561|PI4KB_RAT Phosphatidylinositol 4-kinase beta (PtdIns 4-kinase) (PI4Kbeta) (PI4K-beta) Length = 816 Score = 187 bits (474), Expect = 1e-46 Identities = 106/278 (38%), Positives = 164/278 (58%), Gaps = 3/278 (1%) Frame = +3 Query: 753 EIKENRKNNPIDNKDDIYYQACIFKVGDDVRQDILALQVIRLFKNILEREGLDHYLYPYR 932 +++ R+ +P + + + I K GDD+RQ++LA QV++ ++I E+E + ++ PY+ Sbjct: 540 KVRRIREGSPYGHLPNWRLLSVIVKCGDDLRQELLAFQVLKQLQSIWEQERVPLWIKPYK 599 Query: 933 VVATSPGCGVIECVPDSKSRDQIGRTTDSGMYDYFLSLFGEETTPSYQEARRNFVMSMAS 1112 ++ S G+IE V ++ S Q+ + + + DYFL G TT ++ A+RNFV S A Sbjct: 600 ILVISADSGMIEPVVNAVSIHQVKKQSQLSLLDYFLQEHGSYTTEAFLSAQRNFVQSCAG 659 Query: 1113 YSVVCYLLQIKDRHNGNIMIDKHGHIIHIDFGFMFESSPGNNMGFEPD-IKLTKEMMMVM 1289 Y +VCYLLQ+KDRHNGNI++D GHIIHIDFGF+ SSP N+GFE KLT E + VM Sbjct: 660 YCLVCYLLQVKDRHNGNILLDAEGHIIHIDFGFILSSSP-RNLGFETSAFKLTTEFVDVM 718 Query: 1290 GGKLDSPAFQWFEQLSVQAYLVLRPYQEALVALVSLMLD-SGLPCFRG-QTIKLLRQRFT 1463 GG L+ F +++ L +Q + R + + +V +V +M S LPCF G TI+ L++RF Sbjct: 719 GG-LNGDMFNYYKMLMLQGLIAARKHMDKVVQIVEIMQQGSQLPCFHGSSTIRNLKERFH 777 Query: 1464 PLMSDRDAANHYLRVLRTCCDHWRANTYDFIQYFQNQI 1577 M++ +++ YD QY N I Sbjct: 778 MSMTEEQLQLLVEQMVDGSMRSITTKLYDGFQYLTNGI 815
>sp|Q8BKC8|PI4KB_MOUSE Phosphatidylinositol 4-kinase beta (PtdIns 4-kinase) (PI4Kbeta) (PI4K-beta) Length = 801 Score = 187 bits (474), Expect = 1e-46 Identities = 106/278 (38%), Positives = 164/278 (58%), Gaps = 3/278 (1%) Frame = +3 Query: 753 EIKENRKNNPIDNKDDIYYQACIFKVGDDVRQDILALQVIRLFKNILEREGLDHYLYPYR 932 +++ R+ +P + + + I K GDD+RQ++LA QV++ ++I E+E + ++ PY+ Sbjct: 525 KVRRIREGSPYGHLPNWRLLSVIVKCGDDLRQELLAFQVLKQLQSIWEQERVPLWIKPYK 584 Query: 933 VVATSPGCGVIECVPDSKSRDQIGRTTDSGMYDYFLSLFGEETTPSYQEARRNFVMSMAS 1112 ++ S G+IE V ++ S Q+ + + + DYFL G TT ++ A+RNFV S A Sbjct: 585 ILVISADSGMIEPVVNAVSIHQVKKQSQLSLLDYFLQEHGSYTTEAFLSAQRNFVQSCAG 644 Query: 1113 YSVVCYLLQIKDRHNGNIMIDKHGHIIHIDFGFMFESSPGNNMGFEPD-IKLTKEMMMVM 1289 Y +VCYLLQ+KDRHNGNI++D GHIIHIDFGF+ SSP N+GFE KLT E + VM Sbjct: 645 YCLVCYLLQVKDRHNGNILLDAEGHIIHIDFGFILSSSP-RNLGFETSAFKLTTEFVDVM 703 Query: 1290 GGKLDSPAFQWFEQLSVQAYLVLRPYQEALVALVSLMLD-SGLPCFRG-QTIKLLRQRFT 1463 GG L+ F +++ L +Q + R + + +V +V +M S LPCF G TI+ L++RF Sbjct: 704 GG-LNGDMFNYYKMLMLQGLIAARKHMDKVVQIVEIMQQGSQLPCFHGSSTIRNLKERFH 762 Query: 1464 PLMSDRDAANHYLRVLRTCCDHWRANTYDFIQYFQNQI 1577 M++ +++ YD QY N I Sbjct: 763 MSMTEEQLQLLVEQMVDGSMRSITTKLYDGFQYLTNGI 800
>sp|P54677|PI4K_DICDI Phosphatidylinositol 4-kinase (PI4-kinase) (PtdIns-4-kinase) (PI4K-alpha) Length = 1093 Score = 186 bits (472), Expect = 2e-46 Identities = 103/280 (36%), Positives = 169/280 (60%), Gaps = 5/280 (1%) Frame = +3 Query: 753 EIKENRKNNPIDNKDDIYYQACIFKVGDDVRQDILALQVIRLFKNILEREGLDHYLYPYR 932 +I+ +K +P + + + I K GDD RQ+ +A+Q+I F I + L YL PY Sbjct: 813 KIERYKKISPFGDYPNWRLYSVIVKTGDDCRQEQMAVQLISKFDEIWKETRLPLYLRPYS 872 Query: 933 VVATSPGCGVIECVPDSKSRDQIGRTTD--SGMYDYFLSLFGEETTPSYQEARRNFVMSM 1106 ++ TS G G+IE +PD+ S + ++T + + +YF S +G+ + ++ A+ NF+ SM Sbjct: 873 ILVTSSGGGIIETIPDTMSLHNLKKSTPGFTTLLNYFKSTYGDPSGLRFRTAQSNFIESM 932 Query: 1107 ASYSVVCYLLQIKDRHNGNIMIDKHGHIIHIDFGFMFESSPGNNMGFEPDIKLTKEMMMV 1286 A+YS+V Y+LQIKDRHNGNI+IDK GHI+HIDFGF+ +SPGN KLT+E++ V Sbjct: 933 AAYSIVTYILQIKDRHNGNILIDKEGHIVHIDFGFILSNSPGNISFESAPFKLTQELVDV 992 Query: 1287 MGGKLDSPAFQWFEQLSVQAYLVLRPYQEALVALVSLMLDS-GLPCFRG--QTIKLLRQR 1457 MGG + S FQ+F+ L V+ + R + +++L+ +M+ + CF G + I+ L+ R Sbjct: 993 MGG-IQSGQFQYFKVLCVRGLIEARKQVDKIISLIEIMMSGPKMSCFVGGKEVIEQLKAR 1051 Query: 1458 FTPLMSDRDAANHYLRVLRTCCDHWRANTYDFIQYFQNQI 1577 F +++R+ + ++ DH++ YD Q + N I Sbjct: 1052 FFLDVNERECSTLVENLISYSIDHFKTRYYDKYQSWLNGI 1091
>sp|O02810|PI4KB_BOVIN Phosphatidylinositol 4-kinase beta (PtdIns 4-kinase) (PI4Kbeta) (PI4K-beta) Length = 816 Score = 186 bits (472), Expect = 2e-46 Identities = 106/278 (38%), Positives = 163/278 (58%), Gaps = 3/278 (1%) Frame = +3 Query: 753 EIKENRKNNPIDNKDDIYYQACIFKVGDDVRQDILALQVIRLFKNILEREGLDHYLYPYR 932 +++ R+ +P + + + I K GDD+RQ++LA QV++ ++I E+E + ++ PY+ Sbjct: 540 KVRRIREGSPYGHLPNWRLLSVIVKCGDDLRQELLAFQVLKQLQSIWEQERVPLWIKPYK 599 Query: 933 VVATSPGCGVIECVPDSKSRDQIGRTTDSGMYDYFLSLFGEETTPSYQEARRNFVMSMAS 1112 ++ S G+IE V ++ S Q+ + + + DYFL G TT ++ A+RNFV S A Sbjct: 600 ILVISADSGMIEPVVNAVSIHQVKKQSQLSLLDYFLQEHGSYTTEAFLSAQRNFVQSCAG 659 Query: 1113 YSVVCYLLQIKDRHNGNIMIDKHGHIIHIDFGFMFESSPGNNMGFEPD-IKLTKEMMMVM 1289 Y + CYLLQ+KDRHNGNI++D GHIIHIDFGF+ SSP N+GFE KLT E + VM Sbjct: 660 YCLGCYLLQVKDRHNGNILLDAEGHIIHIDFGFILSSSP-RNLGFETSAFKLTTEFVDVM 718 Query: 1290 GGKLDSPAFQWFEQLSVQAYLVLRPYQEALVALVSLMLD-SGLPCFRG-QTIKLLRQRFT 1463 GG LD F +++ L +Q + R + + +V +V +M S LPCF G TI+ L++RF Sbjct: 719 GG-LDGDMFNYYKMLMLQGLIAARKHMDKVVQIVEIMQQGSQLPCFHGSSTIRNLKERFH 777 Query: 1464 PLMSDRDAANHYLRVLRTCCDHWRANTYDFIQYFQNQI 1577 M++ +++ YD QY N I Sbjct: 778 MSMTEEQLQLLVEQMVDGSMRSITTKLYDGFQYLTNGI 815
>sp|Q9UW24|PIK1A_CANAL Phosphatidylinositol 4-kinase PIK1a (PI4-kinase) (PtdIns-4-kinase) Length = 956 Score = 158 bits (400), Expect = 4e-38 Identities = 99/271 (36%), Positives = 154/271 (56%), Gaps = 18/271 (6%) Frame = +3 Query: 726 ANNFKKQTHEIKENRKNNPIDNKDDIYYQACIFKVGDDVRQDILALQVIRLFKNILEREG 905 A ++ ++ H I+++ ++N D + I K GDD+ Q+ A Q+I L NI ++ Sbjct: 657 AEDWNEKKHRIRKSSIYGHLENWD---LCSVIVKNGDDLPQEAFACQLISLISNIWKKHK 713 Query: 906 LDHYLYPYRVVATSPGCGVIECVPDSKSRDQIGRTT-----DSG---------MYDYFLS 1043 +D + +++ TS G++E + ++ S I ++ ++G + DYF Sbjct: 714 VDFWTKRMKILITSANAGLVETITNAMSIHSIKKSLTELSIENGENAKGRIFTLKDYFQK 773 Query: 1044 LFGEETTPSYQEARRNFVMSMASYSVVCYLLQIKDRHNGNIMIDKHGHIIHIDFGFMFES 1223 L+G + Y +A+ NF +S+ASYS++CY+LQIKDRHNGNIM+D GHIIHIDFGF+ + Sbjct: 774 LYGSVDSSKYVKAQENFAISLASYSIICYVLQIKDRHNGNIMLDHEGHIIHIDFGFLLSN 833 Query: 1224 SPGNNMGFE-PDIKLTKEMMMVMGGKLDSPAFQWFEQLSVQAYLVLRPYQEALVALVSLM 1400 SPG ++GFE KLT E + V+GG L+S A+ F + LR +V++V LM Sbjct: 834 SPG-SVGFEAAPFKLTSEYVEVLGG-LESKAYLKFVDTCKNCFKALRKEWGQIVSIVELM 891 Query: 1401 -LDSGLPCFRG--QTIKLLRQRFTPLMSDRD 1484 S LPCF T LL+QR +SD + Sbjct: 892 QKGSSLPCFNNGDNTSVLLQQRLQLHLSDEE 922
>sp|Q9UW20|PIK1_CANAL Phosphatidylinositol 4-kinase PIK1alpha (PI4-kinase) (PtdIns-4-kinase) Length = 977 Score = 158 bits (400), Expect = 4e-38 Identities = 102/269 (37%), Positives = 153/269 (56%), Gaps = 18/269 (6%) Frame = +3 Query: 732 NFKKQTHEIKENRKNNPIDNKDDIYYQACIFKVGDDVRQDILALQVIRLFKNILEREGLD 911 N KKQ IK++ + N D + I K GDD+ Q+ A Q+I + NI ++ + Sbjct: 682 NTKKQ--RIKKSSAYGHLKNWD---LCSVIAKNGDDLPQEAFACQLISMISNIWKKNNIP 736 Query: 912 HYLYPYRVVATSPGCGVIECVPDSKSRDQIGRT-----TDSG---------MYDYFLSLF 1049 + +++ TS G++E + ++ S I ++ SG + DYF S+F Sbjct: 737 VWTKRMKILITSANTGLVETITNAMSIHSIKKSFTEHSIKSGENSKGKIFTLLDYFHSVF 796 Query: 1050 GEETTPSYQEARRNFVMSMASYSVVCYLLQIKDRHNGNIMIDKHGHIIHIDFGFMFESSP 1229 G + S++ A++NF S+A+YS++CY+LQIKDRHNGNIM+D GHIIHIDFGF+ +SP Sbjct: 797 GSPNSTSFRTAQQNFAKSLAAYSIICYVLQIKDRHNGNIMVDGDGHIIHIDFGFLLSNSP 856 Query: 1230 GNNMGFE-PDIKLTKEMMMVMGGKLDSPAFQWFEQLSVQAYLVLRPYQEALVALVSLM-L 1403 G ++GFE KLT E + ++GG +DS + F L Q + LR E ++ +V LM Sbjct: 857 G-SVGFEAAPFKLTVEYVELLGG-VDSEIYSQFVYLCKQCFKSLRDNSEEIIEIVELMQK 914 Query: 1404 DSGLPCFRG--QTIKLLRQRFTPLMSDRD 1484 DS LPCF T LL+QR ++D D Sbjct: 915 DSTLPCFNNGENTSVLLKQRLQLQLNDED 943
>sp|P39104|PIK1_YEAST Phosphatidylinositol 4-kinase PIK1 (PI4-kinase) (PtdIns-4-kinase) Length = 1066 Score = 157 bits (398), Expect = 7e-38 Identities = 93/241 (38%), Positives = 141/241 (58%), Gaps = 19/241 (7%) Frame = +3 Query: 819 IFKVGDDVRQDILALQVIRLFKNILEREGLDHYLYPYRVVATSPGCGVIECVPDSKSRDQ 998 I K GDD+RQ+ A Q+I+ NI +E +D ++ +++ TS G++E + ++ S Sbjct: 794 IAKTGDDLRQEAFAYQMIQAMANIWVKEKVDVWVKRMKILITSANTGLVETITNAMSVHS 853 Query: 999 IGRTTDSGMY---------------DYFLSLFGEETTPSYQEARRNFVMSMASYSVVCYL 1133 I + M D+FL FG Y+ A+ NF S+A+YSV+CYL Sbjct: 854 IKKALTKKMIEDAELDDKGGIASLNDHFLRAFGNPNGFKYRRAQDNFASSLAAYSVICYL 913 Query: 1134 LQIKDRHNGNIMIDKHGHIIHIDFGFMFESSPGNNMGFE-PDIKLTKEMMMVMGGKLDSP 1310 LQ+KDRHNGNIMID GH+ HIDFGFM +SPG ++GFE KLT E + ++GG ++ Sbjct: 914 LQVKDRHNGNIMIDNEGHVSHIDFGFMLSNSPG-SVGFEAAPFKLTYEYIELLGG-VEGE 971 Query: 1311 AFQWFEQLSVQAYLVLRPYQEALVALVSLM-LDSGLPCFRG--QTIKLLRQRFTPLMSDR 1481 AF+ F +L+ ++ LR Y + +V++ +M D+ PCF QT LRQRF +S++ Sbjct: 972 AFKKFVELTKSSFKALRKYADQIVSMCEIMQKDNMQPCFDAGEQTSVQLRQRFQLDLSEK 1031 Query: 1482 D 1484 + Sbjct: 1032 E 1032
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 191,546,594
Number of Sequences: 369166
Number of extensions: 3825499
Number of successful extensions: 9595
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 8940
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 9517
length of database: 68,354,980
effective HSP length: 116
effective length of database: 46,925,720
effective search space used: 22148939840
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail