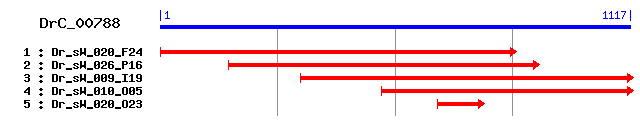
DrC_00788
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00788 (1117 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P49900|ARGI_RANCA Arginase, hepatic 261 3e-69 sp|P78540|ARGI2_HUMAN Arginase-2, mitochondrial precursor (... 245 2e-64 sp|P05089|ARGI1_HUMAN Arginase-1 (Type I arginase) (Liver-t... 244 3e-64 sp|O08691|ARGI2_MOUSE Arginase-2, mitochondrial precursor (... 243 7e-64 sp|Q95JC8|ARGI1_PIG Arginase-1 (Type I arginase) (Liver-typ... 242 2e-63 sp|Q91554|ARGN2_XENLA Arginase, non-hepatic 2 241 3e-63 sp|O08701|ARGI2_RAT Arginase-2, mitochondrial precursor (Ar... 241 4e-63 sp|Q61176|ARGI1_MOUSE Arginase-1 (Type I arginase) (Liver-t... 239 8e-63 sp|Q91555|ARGN3_XENLA Arginase, non-hepatic 3 239 8e-63 sp|Q91553|ARGN1_XENLA Arginase, non-hepatic 1 239 1e-62
>sp|P49900|ARGI_RANCA Arginase, hepatic Length = 323 Score = 261 bits (666), Expect = 3e-69 Identities = 132/267 (49%), Positives = 177/267 (66%), Gaps = 1/267 (0%) Frame = +3 Query: 162 DLEFHDFGNIDTNLIGNDNNIFNAKYPRMFLYNTMKIADKIEEMIDKNYIPLTIGGDHSI 341 + E D+G++ + D N K PR K+A+ + E+ + LT+GGDHS+ Sbjct: 45 EYEVRDYGDLHFPELPCDEPFQNVKNPRTVGQAAEKVANAVSEVKRSGRVCLTLGGDHSL 104 Query: 342 TTGTILGHARRHEDLCVIWVDAHADINTPLSSSSGNMHGMPLSFLIHEMQNEMPWIEEFK 521 GTI GHA+ H DLCV+WVDAHADINTP++S SGN+HG P+SFLI E+Q ++P I F Sbjct: 105 AVGTITGHAKVHPDLCVVWVDAHADINTPITSPSGNLHGQPVSFLIRELQTKVPAIPGFS 164 Query: 522 DIPPVISAKNIVYIGLRSVDPYERHVLNKYNIKSFSMEDIDRDGINCVIQQAIKAVNPNL 701 + P +SAK+IVYIGLR VDP E ++L IKS+SM D+DR IN V+++ I+ + Sbjct: 165 WVQPSLSAKDIVYIGLRDVDPGEHYILKTLGIKSYSMSDVDRLTINKVMEETIEFLVGKK 224 Query: 702 DRYFHLSFDIDALDPLYAPSTGTAVPGGLTLREGVYLCEQLFNTGKLAAIDVTEINTQVG 881 R HLSFDID LDP AP+TGT VPGGLT REG+Y+ EQL+NTG L+A+D+ E+N G Sbjct: 225 KRPIHLSFDIDGLDPSVAPATGTPVPGGLTYREGMYITEQLYNTGLLSAVDMMEVNPSRG 284 Query: 882 -TYSEVVTTINSATKIIQGCLGIKREG 959 T E T+N++ +I C G REG Sbjct: 285 ETERESKLTVNTSLNMILSCFGKAREG 311
Score = 33.9 bits (76), Expect = 0.87
Identities = 14/33 (42%), Positives = 22/33 (66%)
Frame = +2
Query: 2 LLGVPFEKGQSLSGVSCGPKLIRTKKIVQQIED 100
+LG PF KGQ+ GV GP IR +++++E+
Sbjct: 11 VLGAPFSKGQARGGVEEGPIYIRRAGLIEKLEE 43
>sp|P78540|ARGI2_HUMAN Arginase-2, mitochondrial precursor (Arginase II) (Non-hepatic arginase) (Kidney-type arginase) Length = 354 Score = 245 bits (625), Expect = 2e-64 Identities = 129/276 (46%), Positives = 175/276 (63%), Gaps = 4/276 (1%) Frame = +3 Query: 156 ISDLEFH--DFGNIDTNLIGNDNNIFNAKY-PRMFLYNTMKIADKIEEMIDKNYIPLTIG 326 +S L H DFG++ + D+ N PR ++A+ + + Y +T+G Sbjct: 58 LSSLGCHLKDFGDLSFTPVPKDDLYNNLIVNPRSVGLANQELAEVVSRAVSDGYSCVTLG 117 Query: 327 GDHSITTGTILGHARRHEDLCVIWVDAHADINTPLSSSSGNMHGMPLSFLIHEMQNEMPW 506 GDHS+ GTI GHAR DLCV+WVDAHADINTPL++SSGN+HG P+SFL+ E+Q+++P Sbjct: 118 GDHSLAIGTISGHARHCPDLCVVWVDAHADINTPLTTSSGNLHGQPVSFLLRELQDKVPQ 177 Query: 507 IEEFKDIPPVISAKNIVYIGLRSVDPYERHVLNKYNIKSFSMEDIDRDGINCVIQQAIKA 686 + F I P IS+ +IVYIGLR VDP E +L Y+I+ FSM DIDR GI V+++ Sbjct: 178 LPGFSWIKPCISSASIVYIGLRDVDPPEHFILKNYDIQYFSMRDIDRLGIQKVMERTFDL 237 Query: 687 VNPNLDRYFHLSFDIDALDPLYAPSTGTAVPGGLTLREGVYLCEQLFNTGKLAAIDVTEI 866 + R HLSFDIDA DP AP+TGT V GGLT REG+Y+ E++ NTG L+A+D+ E+ Sbjct: 238 LIGKRQRPIHLSFDIDAFDPTLAPATGTPVVGGLTYREGMYIAEEIHNTGLLSALDLVEV 297 Query: 867 NTQVGT-YSEVVTTINSATKIIQGCLGIKREGAPLI 971 N Q+ T E TT N A +I G REG ++ Sbjct: 298 NPQLATSEEEAKTTANLAVDVIASSFGQTREGGHIV 333
>sp|P05089|ARGI1_HUMAN Arginase-1 (Type I arginase) (Liver-type arginase) Length = 322 Score = 244 bits (623), Expect = 3e-64 Identities = 122/267 (45%), Positives = 174/267 (65%), Gaps = 1/267 (0%) Frame = +3 Query: 162 DLEFHDFGNIDTNLIGNDNNIFNAKYPRMFLYNTMKIADKIEEMIDKNYIPLTIGGDHSI 341 + + D+G++ I ND+ K PR + ++A K+ E+ I L +GGDHS+ Sbjct: 44 ECDVKDYGDLPFADIPNDSPFQIVKNPRSVGKASEQLAGKVAEVKKNGRISLVLGGDHSL 103 Query: 342 TTGTILGHARRHEDLCVIWVDAHADINTPLSSSSGNMHGMPLSFLIHEMQNEMPWIEEFK 521 G+I GHAR H DL VIWVDAH DINTPL+++SGN+HG P+SFL+ E++ ++P + F Sbjct: 104 AIGSISGHARVHPDLGVIWVDAHTDINTPLTTTSGNLHGQPVSFLLKELKGKIPDVPGFS 163 Query: 522 DIPPVISAKNIVYIGLRSVDPYERHVLNKYNIKSFSMEDIDRDGINCVIQQAIKAVNPNL 701 + P ISAK+IVYIGLR VDP E ++L IK FSM ++DR GI V+++ + + Sbjct: 164 WVTPCISAKDIVYIGLRDVDPGEHYILKTLGIKYFSMTEVDRLGIGKVMEETLSYLLGRK 223 Query: 702 DRYFHLSFDIDALDPLYAPSTGTAVPGGLTLREGVYLCEQLFNTGKLAAIDVTEINTQVG 881 R HLSFD+D LDP + P+TGT V GGLT REG+Y+ E+++ TG L+ +D+ E+N +G Sbjct: 224 KRPIHLSFDVDGLDPSFTPATGTPVVGGLTYREGLYITEEIYKTGLLSGLDIMEVNPSLG 283 Query: 882 -TYSEVVTTINSATKIIQGCLGIKREG 959 T EV T+N+A I C G+ REG Sbjct: 284 KTPEEVTRTVNTAVAITLACFGLAREG 310
Score = 31.2 bits (69), Expect = 5.6
Identities = 11/33 (33%), Positives = 22/33 (66%)
Frame = +2
Query: 2 LLGVPFEKGQSLSGVSCGPKLIRTKKIVQQIED 100
++G PF KGQ GV GP ++R +++++++
Sbjct: 10 IIGAPFSKGQPRGGVEEGPTVLRKAGLLEKLKE 42
>sp|O08691|ARGI2_MOUSE Arginase-2, mitochondrial precursor (Arginase II) (Non-hepatic arginase) (Kidney-type arginase) Length = 354 Score = 243 bits (620), Expect = 7e-64 Identities = 126/267 (47%), Positives = 168/267 (62%), Gaps = 2/267 (0%) Frame = +3 Query: 177 DFGNIDTNLIGNDNNIFN-AKYPRMFLYNTMKIADKIEEMIDKNYIPLTIGGDHSITTGT 353 DFG++ + D+ N YPR ++A+ + + Y +T+GGDHS+ GT Sbjct: 67 DFGDLSFTNVPQDDPYNNLVVYPRSVGLANQELAEVVSRAVSGGYSCVTMGGDHSLAIGT 126 Query: 354 ILGHARRHEDLCVIWVDAHADINTPLSSSSGNMHGMPLSFLIHEMQNEMPWIEEFKDIPP 533 I+GHAR DLCVIWVDAHADINTPL++ SGN+HG PLSFLI E+Q+++P + F I P Sbjct: 127 IIGHARHRPDLCVIWVDAHADINTPLTTVSGNIHGQPLSFLIKELQDKVPQLPGFSWIKP 186 Query: 534 VISAKNIVYIGLRSVDPYERHVLNKYNIKSFSMEDIDRDGINCVIQQAIKAVNPNLDRYF 713 +S NIVYIGLR V+P E +L Y+I+ FSM +IDR GI V++Q + R Sbjct: 187 CLSPPNIVYIGLRDVEPPEHFILKNYDIQYFSMREIDRLGIQKVMEQTFDRLIGKRQRPI 246 Query: 714 HLSFDIDALDPLYAPSTGTAVPGGLTLREGVYLCEQLFNTGKLAAIDVTEINTQVGT-YS 890 HLSFDIDA DP AP+TGT V GGLT REGVY+ E++ NTG L+A+D+ E+N + T Sbjct: 247 HLSFDIDAFDPKLAPATGTPVVGGLTYREGVYITEEIHNTGLLSALDLVEVNPHLATSEE 306 Query: 891 EVVTTINSATKIIQGCLGIKREGAPLI 971 E T A +I G REG ++ Sbjct: 307 EAKATARLAVDVIASSFGQTREGGHIV 333
>sp|Q95JC8|ARGI1_PIG Arginase-1 (Type I arginase) (Liver-type arginase) Length = 322 Score = 242 bits (617), Expect = 2e-63 Identities = 116/267 (43%), Positives = 172/267 (64%), Gaps = 1/267 (0%) Frame = +3 Query: 162 DLEFHDFGNIDTNLIGNDNNIFNAKYPRMFLYNTMKIADKIEEMIDKNYIPLTIGGDHSI 341 + + D+G++ + ND K PR ++AD + E+ L +GGDHS+ Sbjct: 44 ECDVKDYGDLCFADVPNDTPFQIVKNPRSVGKANQQLADVVAEIKKNGRTSLVLGGDHSM 103 Query: 342 TTGTILGHARRHEDLCVIWVDAHADINTPLSSSSGNMHGMPLSFLIHEMQNEMPWIEEFK 521 G+I GHAR H DLCVIWVDAH DINTPL++++GN+HG P+SFL+ E++ ++P + Sbjct: 104 AIGSISGHARVHPDLCVIWVDAHTDINTPLTTTTGNLHGQPVSFLLKELKEKIPEVPGLS 163 Query: 522 DIPPVISAKNIVYIGLRSVDPYERHVLNKYNIKSFSMEDIDRDGINCVIQQAIKAVNPNL 701 + P +SAK+IVYIGLR VDP E ++L IK FSM ++D+ GI V+++A + Sbjct: 164 WVTPCLSAKDIVYIGLRDVDPAEHYILKTLGIKYFSMIEVDKLGIGKVMEEAFSYLLGRK 223 Query: 702 DRYFHLSFDIDALDPLYAPSTGTAVPGGLTLREGVYLCEQLFNTGKLAAIDVTEINTQVG 881 R HLSFD+D LDP + P+TGT V GGL+ REG+Y+ E+++ TG L+ +D+ E+N +G Sbjct: 224 KRPIHLSFDVDGLDPFFTPATGTPVHGGLSYREGIYITEEIYKTGLLSGLDIMEVNPSLG 283 Query: 882 -TYSEVVTTINSATKIIQGCLGIKREG 959 T EV T+N+A ++ C G+ REG Sbjct: 284 KTPEEVTRTVNTAVALVLACFGVAREG 310
Score = 30.4 bits (67), Expect = 9.6
Identities = 11/33 (33%), Positives = 21/33 (63%)
Frame = +2
Query: 2 LLGVPFEKGQSLSGVSCGPKLIRTKKIVQQIED 100
++G PF KGQ GV GP +R +++++++
Sbjct: 10 IIGAPFSKGQPRGGVEEGPTALRKAGLLEKLKE 42
>sp|Q91554|ARGN2_XENLA Arginase, non-hepatic 2 Length = 360 Score = 241 bits (615), Expect = 3e-63 Identities = 124/265 (46%), Positives = 173/265 (65%), Gaps = 3/265 (1%) Frame = +3 Query: 177 DFGNIDTNLIGNDNNIFNA--KYPRMFLYNTMKIADKIEEMIDKNYIPLTIGGDHSITTG 350 DFG++ + + ND ++N+ K+PR +A+++ + + + +T+GGDHS+ G Sbjct: 69 DFGDLHFSKVPNDE-LYNSIVKHPRTVGLACKVLAEEVSKAVGAGHTCVTLGGDHSLAFG 127 Query: 351 TILGHARRHEDLCVIWVDAHADINTPLSSSSGNMHGMPLSFLIHEMQNEMPWIEEFKDIP 530 +I GHA++ DLCVIWVDAHADINTPL++ SGN+HG P+SFL+ E+Q+++P I F Sbjct: 128 SITGHAQQCPDLCVIWVDAHADINTPLTTPSGNLHGQPVSFLLRELQDKVPPIPGFSWAK 187 Query: 531 PVISAKNIVYIGLRSVDPYERHVLNKYNIKSFSMEDIDRDGINCVIQQAIKAVNPNLDRY 710 P +S +IVYIGLR +DP E+ +L YNI +SM ID GI V+++ + DR Sbjct: 188 PCLSKSDIVYIGLRDLDPAEQFILKNYNISYYSMRHIDCMGIKKVMEKTFDQLLGRRDRP 247 Query: 711 FHLSFDIDALDPLYAPSTGTAVPGGLTLREGVYLCEQLFNTGKLAAIDVTEIN-TQVGTY 887 HLSFDIDA DP AP+TGT V GGLT REGVY+ E++ NTG L+A+D+ E+N T Sbjct: 248 IHLSFDIDAFDPALAPATGTPVIGGLTYREGVYITEEIHNTGMLSAVDLVEVNPVLAATS 307 Query: 888 SEVVTTINSATKIIQGCLGIKREGA 962 EV T N A +I C G REGA Sbjct: 308 EEVKATANLAVDVIASCFGQTREGA 332
Score = 32.0 bits (71), Expect = 3.3
Identities = 12/33 (36%), Positives = 21/33 (63%)
Frame = +2
Query: 2 LLGVPFEKGQSLSGVSCGPKLIRTKKIVQQIED 100
++G PF KGQ GV GP IR+ +++++ +
Sbjct: 30 VIGAPFSKGQKRRGVEHGPAAIRSAGLIERLSN 62
>sp|O08701|ARGI2_RAT Arginase-2, mitochondrial precursor (Arginase II) (Non-hepatic arginase) (Kidney-type arginase) Length = 354 Score = 241 bits (614), Expect = 4e-63 Identities = 124/263 (47%), Positives = 167/263 (63%), Gaps = 2/263 (0%) Frame = +3 Query: 177 DFGNIDTNLIGNDNNIFN-AKYPRMFLYNTMKIADKIEEMIDKNYIPLTIGGDHSITTGT 353 DFG++ + D+ N YPR ++A+ + + Y +T+GGDHS+ GT Sbjct: 67 DFGDLSFTNVPKDDPYNNLVVYPRSVGIANQELAEVVSRAVSGGYSCVTLGGDHSLAIGT 126 Query: 354 ILGHARRHEDLCVIWVDAHADINTPLSSSSGNMHGMPLSFLIHEMQNEMPWIEEFKDIPP 533 I GHAR H DLCVIWVDAHADINTPL++ SGN+HG PLSFLI E+Q+++P + F I P Sbjct: 127 ISGHARHHPDLCVIWVDAHADINTPLTTVSGNIHGQPLSFLIRELQDKVPQLPGFSWIKP 186 Query: 534 VISAKNIVYIGLRSVDPYERHVLNKYNIKSFSMEDIDRDGINCVIQQAIKAVNPNLDRYF 713 +S N+VYIGLR V+P E +L ++I+ FSM DIDR GI V++Q + R Sbjct: 187 CLSPPNLVYIGLRDVEPAEHFILKSFDIQYFSMRDIDRLGIQKVMEQTFDRLIGKRKRPI 246 Query: 714 HLSFDIDALDPLYAPSTGTAVPGGLTLREGVYLCEQLFNTGKLAAIDVTEINTQVGT-YS 890 HLSFDIDA DP AP+TGT V GGLT REG+Y+ E++ +TG L+A+D+ E+N + T Sbjct: 247 HLSFDIDAFDPKLAPATGTPVVGGLTYREGLYITEEIHSTGLLSALDLVEVNPHLATSEE 306 Query: 891 EVVTTINSATKIIQGCLGIKREG 959 E T + A +I G REG Sbjct: 307 EAKATASLAVDVIASSFGQTREG 329
>sp|Q61176|ARGI1_MOUSE Arginase-1 (Type I arginase) (Liver-type arginase) Length = 323 Score = 239 bits (611), Expect = 8e-63 Identities = 118/276 (42%), Positives = 175/276 (63%), Gaps = 1/276 (0%) Frame = +3 Query: 159 SDLEFHDFGNIDTNLIGNDNNIFNAKYPRMFLYNTMKIADKIEEMIDKNYIPLTIGGDHS 338 ++ + D G++ + ND++ K PR ++A + E+ + + +GGDHS Sbjct: 43 TEYDVRDHGDLAFVDVPNDSSFQIVKNPRSVGKANEELAGVVAEVQKNGRVSVVLGGDHS 102 Query: 339 ITTGTILGHARRHEDLCVIWVDAHADINTPLSSSSGNMHGMPLSFLIHEMQNEMPWIEEF 518 + G+I GHAR H DLCVIWVDAH DINTPL++SSGN+HG P+SFL+ E++ + P + F Sbjct: 103 LAVGSISGHARVHPDLCVIWVDAHTDINTPLTTSSGNLHGQPVSFLLKELKGKFPDVPGF 162 Query: 519 KDIPPVISAKNIVYIGLRSVDPYERHVLNKYNIKSFSMEDIDRDGINCVIQQAIKAVNPN 698 + P ISAK+IVYIGLR VDP E +++ IK FSM ++D+ GI V+++ + Sbjct: 163 SWVTPCISAKDIVYIGLRDVDPGEHYIIKTLGIKYFSMTEVDKLGIGKVMEETFSYLLGR 222 Query: 699 LDRYFHLSFDIDALDPLYAPSTGTAVPGGLTLREGVYLCEQLFNTGKLAAIDVTEINTQV 878 R HLSFD+D LDP + P+TGT V GGL+ REG+Y+ E+++ TG L+ +D+ E+N + Sbjct: 223 KKRPIHLSFDVDGLDPAFTPATGTPVLGGLSYREGLYITEEIYKTGLLSGLDIMEVNPTL 282 Query: 879 G-TYSEVVTTINSATKIIQGCLGIKREGAPLITPDY 983 G T EV +T+N+A + C G +REG DY Sbjct: 283 GKTAEEVKSTVNTAVALTLACFGTQREGNHKPGTDY 318
Score = 30.8 bits (68), Expect = 7.3
Identities = 11/33 (33%), Positives = 21/33 (63%)
Frame = +2
Query: 2 LLGVPFEKGQSLSGVSCGPKLIRTKKIVQQIED 100
++G PF KGQ GV GP +R +++++++
Sbjct: 10 IIGAPFSKGQPRGGVEKGPAALRKAGLLEKLKE 42
>sp|Q91555|ARGN3_XENLA Arginase, non-hepatic 3 Length = 360 Score = 239 bits (611), Expect = 8e-63 Identities = 124/264 (46%), Positives = 171/264 (64%), Gaps = 2/264 (0%) Frame = +3 Query: 177 DFGNIDTNLIGNDNNIFN-AKYPRMFLYNTMKIADKIEEMIDKNYIPLTIGGDHSITTGT 353 DFG++ + + ND + K+PR +A ++ + + + +T+GGDHS+ G+ Sbjct: 69 DFGDLHFSQVPNDEQYNSIVKHPRTVGLACKVLAKEVGKAVGAGHTCVTLGGDHSLAFGS 128 Query: 354 ILGHARRHEDLCVIWVDAHADINTPLSSSSGNMHGMPLSFLIHEMQNEMPWIEEFKDIPP 533 I GHA++ DLCVIWVDAHADINTPL++ SGN+HG P+SFL+ E+Q+++P I F P Sbjct: 129 ITGHAQQCPDLCVIWVDAHADINTPLTTPSGNLHGQPVSFLLRELQDKIPPIPGFSWAKP 188 Query: 534 VISAKNIVYIGLRSVDPYERHVLNKYNIKSFSMEDIDRDGINCVIQQAIKAVNPNLDRYF 713 +S +IVYIGLR +DP E+ +L YNI +SM ID GI V+++ + DR Sbjct: 189 CLSKSDIVYIGLRDLDPAEQFILKNYNISYYSMRHIDCMGIRKVMEKTFDQLLGRRDRPI 248 Query: 714 HLSFDIDALDPLYAPSTGTAVPGGLTLREGVYLCEQLFNTGKLAAIDVTEINTQVGTYS- 890 HLSFDIDA DP AP+TGT V GGLT REGVY+ E++ NTG L+A+D+ E+N + T S Sbjct: 249 HLSFDIDAFDPALAPATGTPVIGGLTYREGVYITEEIHNTGMLSALDLVEVNPVLATTSE 308 Query: 891 EVVTTINSATKIIQGCLGIKREGA 962 EV T N A +I C G REGA Sbjct: 309 EVKATANLAVDVIASCFGQTREGA 332
Score = 31.2 bits (69), Expect = 5.6
Identities = 12/33 (36%), Positives = 20/33 (60%)
Frame = +2
Query: 2 LLGVPFEKGQSLSGVSCGPKLIRTKKIVQQIED 100
++G PF KGQ GV GP IR+ ++ ++ +
Sbjct: 30 VIGAPFSKGQKRRGVEHGPAAIRSAGLIDRLSN 62
>sp|Q91553|ARGN1_XENLA Arginase, non-hepatic 1 Length = 360 Score = 239 bits (610), Expect = 1e-62 Identities = 123/265 (46%), Positives = 173/265 (65%), Gaps = 3/265 (1%) Frame = +3 Query: 177 DFGNIDTNLIGNDNNIFNA--KYPRMFLYNTMKIADKIEEMIDKNYIPLTIGGDHSITTG 350 DFG++ + + ND ++N+ K+PR +A+++ + + + +T+GGDHS+ G Sbjct: 69 DFGDLHFSQVPNDE-LYNSIVKHPRTVGLACKVLAEEVSKAVGAGHTCVTLGGDHSLAFG 127 Query: 351 TILGHARRHEDLCVIWVDAHADINTPLSSSSGNMHGMPLSFLIHEMQNEMPWIEEFKDIP 530 +I GHA++ DLCVIWVDAHADINTPL++ SGN+HG P+SFL+ E+Q+++P I F Sbjct: 128 SITGHAQQCPDLCVIWVDAHADINTPLTTPSGNLHGQPVSFLLRELQDKVPPIPGFSWAK 187 Query: 531 PVISAKNIVYIGLRSVDPYERHVLNKYNIKSFSMEDIDRDGINCVIQQAIKAVNPNLDRY 710 P +S +IVYIGLR +DP E+ +L Y+I +SM ID GI V+++ + DR Sbjct: 188 PCLSKSDIVYIGLRDLDPAEQFILKNYDISYYSMRHIDCMGIKKVMEKTFDQLLGRRDRP 247 Query: 711 FHLSFDIDALDPLYAPSTGTAVPGGLTLREGVYLCEQLFNTGKLAAIDVTEIN-TQVGTY 887 HLSFDIDA DP AP+TGT V GGLT REGVY+ E++ NTG L+A+D+ E+N T Sbjct: 248 IHLSFDIDAFDPALAPATGTPVIGGLTYREGVYITEEIHNTGMLSAVDLVEVNPVLAATS 307 Query: 888 SEVVTTINSATKIIQGCLGIKREGA 962 EV T N A +I C G REGA Sbjct: 308 EEVKATANLAVDVIASCFGQTREGA 332
Score = 32.0 bits (71), Expect = 3.3
Identities = 12/33 (36%), Positives = 21/33 (63%)
Frame = +2
Query: 2 LLGVPFEKGQSLSGVSCGPKLIRTKKIVQQIED 100
++G PF KGQ GV GP IR+ +++++ +
Sbjct: 30 VIGAPFSKGQKRRGVEHGPAAIRSAGLIERLSN 62
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 113,544,212
Number of Sequences: 369166
Number of extensions: 2222438
Number of successful extensions: 6040
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 5708
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5966
length of database: 68,354,980
effective HSP length: 112
effective length of database: 47,664,660
effective search space used: 12345146940
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail