DrC_00699
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00699 (1262 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P37276|DYHC_DROME Dynein heavy chain, cytosolic (DYHC) 334 4e-91 sp|Q9JHU4|DYHC_MOUSE Dynein heavy chain, cytosolic (DYHC) (... 329 9e-90 sp|Q14204|DYHC_HUMAN Dynein heavy chain, cytosolic (DYHC) (... 325 2e-88 sp|P38650|DYHC_RAT Dynein heavy chain, cytosolic (DYHC) (Cy... 322 1e-87 sp|Q19020|DYHC_CAEEL Dynein heavy chain, cytosolic (DYHC) 255 2e-67 sp|P34036|DYHC_DICDI Dynein heavy chain, cytosolic (DYHC) 219 1e-56 sp|Q27171|DYHC_PARTE Dynein heavy chain, cytosolic (DYHC) (... 208 3e-53 sp|Q8VHE6|DYH5_MOUSE Ciliary dynein heavy chain 5 (Axonemal... 65 3e-10 sp|Q8TE73|DYH5_HUMAN Ciliary dynein heavy chain 5 (Axonemal... 64 7e-10 sp|P78716|DYHC_FUSSO Dynein heavy chain, cytosolic (DYHC) 64 7e-10
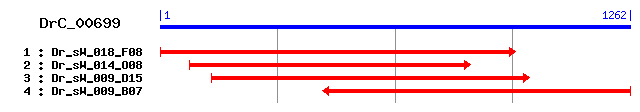
>sp|P37276|DYHC_DROME Dynein heavy chain, cytosolic (DYHC) Length = 4639 Score = 334 bits (856), Expect = 4e-91 Identities = 190/399 (47%), Positives = 247/399 (61%), Gaps = 13/399 (3%) Frame = +1 Query: 1 QRLLNSFLEYLFNEKCFSNNFALTFENDDKT----EIVFPEFNNHGDILQWIYKLPDSQT 168 QRLL SFL+ LF + F +FAL D + I P+ L+WI L D QT Sbjct: 4246 QRLLTSFLKKLFTARSFEADFALVANVDGASGGLRHITMPDGTRRDHFLKWIENLTDRQT 4305 Query: 169 PSWLGLPNSAEKVLLTNLGQSLIGNILKMQQTLEDDEEI---VVDQRKSKRTIELGDSRP 339 PSWLGLPN+AEKVLLT G L+ +LKMQQ LEDD+E+ V DQ + D RP Sbjct: 4306 PSWLGLPNNAEKVLLTTRGTDLVSKLLKMQQ-LEDDDELAYSVEDQSEQSAVGRGEDGRP 4364 Query: 340 VWMRQLYQSTQTWLQLLPKVVIPLQRTTENIKDPIYRCFEREVNSGSNLLLIVRKDLEDI 519 WM+ L+ S WL+LLPK + L+RT ENIKDP+YR FEREV SGS LL V DL+D+ Sbjct: 4365 SWMKTLHNSATAWLELLPKNLQVLKRTVENIKDPLYRYFEREVTSGSRLLQTVILDLQDV 4424 Query: 520 SEVCSGTKKPTNHHRSMMTNLAKGIIPISWIKYPISSGLTVIQWIHDFADRVKQLIHISN 699 +C G KK TNHHRSM++ L +GIIP W +Y + +G TVIQWI DF++RV+QL +S Sbjct: 4425 VLICQGEKKQTNHHRSMLSELVRGIIPKGWKRYTVPAGCTVIQWITDFSNRVQQLQKVSQ 4484 Query: 700 TAINSGAAGLRSVKIWLGGLFMAEAYTTAIRQAVAQSNKWALEELFLDVELGEG--KIDK 873 +GA L+ +WLGGL EAY TA RQ VAQ+N W+LEEL LDV + + K D+ Sbjct: 4485 LVSQAGAKELQGFPVWLGGLLNPEAYITATRQCVAQANSWSLEELALDVTITDAGLKNDQ 4544 Query: 874 NTCI--VTGLRLMGSEIVDSKTLKLSTTIYKDIDICRLKWIRIEKFEHFEENLIRDKVCL 1047 C VTGL+L G++ +++ L L++TI D+ + LKWI+I K+ L Sbjct: 4545 KDCCFGVTGLKLQGAQCKNNELL-LASTIMMDLPVTILKWIKISSEPRI------SKLTL 4597 Query: 1048 PVYLNAARTSILFT--LYFNTTDFINDFYKRGVAILASS 1158 PVYLN+ RT +LFT L + FY+RGVA+L S+ Sbjct: 4598 PVYLNSTRTELLFTVDLAVAAGQESHSFYERGVAVLTST 4636
>sp|Q9JHU4|DYHC_MOUSE Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain) Length = 4644 Score = 329 bits (844), Expect = 9e-90 Identities = 173/390 (44%), Positives = 246/390 (63%), Gaps = 5/390 (1%) Frame = +1 Query: 1 QRLLNSFLEYLFNEKCFSNNFALTFENDDKTEIVFPEFNNHGDILQWIYKLPDSQTPSWL 180 QRLLN+FLE LF + F + F L + D +I P+ + +QW+ LPD+QTPSWL Sbjct: 4260 QRLLNTFLERLFTTRSFDSEFKLACKVDGHKDIQMPDGIRREEFVQWVELLPDAQTPSWL 4319 Query: 181 GLPNSAEKVLLTNLGQSLIGNILKMQQTLEDDEEIVVDQRKSKRTIELGDSRPVWMRQLY 360 GLPN+AE+VLLT G +I +LKMQ ++D+ + K RT D RP WMR L+ Sbjct: 4320 GLPNNAERVLLTTQGVDMISKMLKMQMLEDEDDLAYAETEKKARTDSTSDGRPAWMRTLH 4379 Query: 361 QSTQTWLQLLPKVVIPLQRTTENIKDPIYRCFEREVNSGSNLLLIVRKDLEDISEVCSGT 540 + WL L+P+ + PL+RT ENIKDP++R FEREV G+ LL VR+DL D+ +VC G Sbjct: 4380 TTASNWLHLIPQTLSPLKRTVENIKDPLFRFFEREVKMGAKLLQDVRQDLADVVQVCEGK 4439 Query: 541 KKPTNHHRSMMTNLAKGIIPISWIKYPISSGLTVIQWIHDFADRVKQLIHISNTAINSGA 720 KK TN+ R+++ L KGI+P SW Y + +G+TVIQW+ DF++R+KQL +IS A + GA Sbjct: 4440 KKQTNYLRTLINELVKGILPRSWSHYTVPAGMTVIQWVSDFSERIKQLQNISQAAASGGA 4499 Query: 721 AGLRSVKIWLGGLFMAEAYTTAIRQAVAQSNKWALEELFLDVELGEGK---IDKNTCIVT 891 L+++ + LGGLF+ EAY TA RQ VAQ+N W+LEEL L+V + + +D + VT Sbjct: 4500 KELKNIHVCLGGLFVPEAYITATRQYVAQANSWSLEELCLEVNVTASQSATLDACSFGVT 4559 Query: 892 GLRLMGSEIVDSKTLKLSTTIYKDIDICRLKWIRIEKFEHFEENLIRDKVCLPVYLNAAR 1071 GL+L G+ ++K L LS I + + +L+W++ E V LPVYLN R Sbjct: 4560 GLKLQGATCSNNK-LSLSNAISTVLPLTQLRWVKQTSAEK-----KASVVTLPVYLNFTR 4613 Query: 1072 TSILFTLYFN--TTDFINDFYKRGVAILAS 1155 ++FT+ F T + FY+RGVA+L + Sbjct: 4614 ADLIFTVDFEIATKEDPRSFYERGVAVLCT 4643
>sp|Q14204|DYHC_HUMAN Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain 1) (DHC1) Length = 4646 Score = 325 bits (832), Expect = 2e-88 Identities = 172/390 (44%), Positives = 245/390 (62%), Gaps = 5/390 (1%) Frame = +1 Query: 1 QRLLNSFLEYLFNEKCFSNNFALTFENDDKTEIVFPEFNNHGDILQWIYKLPDSQTPSWL 180 QRLLN+FLE LF + F + F L + D +I P+ + +QW+ LPD+QTPSWL Sbjct: 4262 QRLLNTFLERLFTTRSFDSEFKLACKVDGHKDIQMPDGIRREEFVQWVELLPDTQTPSWL 4321 Query: 181 GLPNSAEKVLLTNLGQSLIGNILKMQQTLEDDEEIVVDQRKSKRTIELGDSRPVWMRQLY 360 GLPN+AE+VLLT G +I +LKMQ ++D+ + K RT D RP WMR L+ Sbjct: 4322 GLPNNAERVLLTTQGVDMISKMLKMQMLEDEDDLAYAETEKKTRTDSTSDGRPAWMRTLH 4381 Query: 361 QSTQTWLQLLPKVVIPLQRTTENIKDPIYRCFEREVNSGSNLLLIVRKDLEDISEVCSGT 540 + WL L+P+ + L+RT ENIKDP++R FEREV G+ LL VR+DL D+ +VC G Sbjct: 4382 TTASNWLHLIPQTLSHLKRTVENIKDPLFRFFEREVKMGAKLLQDVRQDLADVVQVCEGK 4441 Query: 541 KKPTNHHRSMMTNLAKGIIPISWIKYPISSGLTVIQWIHDFADRVKQLIHISNTAINSGA 720 KK TN+ R+++ L KGI+P SW Y + +G+TVIQW+ DF++R+KQL +IS A + GA Sbjct: 4442 KKQTNYLRTLINELVKGILPRSWSHYTVPAGMTVIQWVSDFSERIKQLQNISLAAASGGA 4501 Query: 721 AGLRSVKIWLGGLFMAEAYTTAIRQAVAQSNKWALEELFLDVELGEGK---IDKNTCIVT 891 L+++ + LGGLF+ EAY TA RQ VAQ+N W+LEEL L+V + + +D + VT Sbjct: 4502 KELKNIHVCLGGLFVPEAYITATRQYVAQANSWSLEELCLEVNVTTSQGATLDACSFGVT 4561 Query: 892 GLRLMGSEIVDSKTLKLSTTIYKDIDICRLKWIRIEKFEHFEENLIRDKVCLPVYLNAAR 1071 GL+L G+ ++K L LS I + + +L+W++ E V LPVYLN R Sbjct: 4562 GLKLQGATCNNNK-LSLSNAISTALPLTQLRWVKQTNTEK-----KASVVTLPVYLNFTR 4615 Query: 1072 TSILFTLYFN--TTDFINDFYKRGVAILAS 1155 ++FT+ F T + FY+RGVA+L + Sbjct: 4616 ADLIFTVDFEIATKEDPRSFYERGVAVLCT 4645
>sp|P38650|DYHC_RAT Dynein heavy chain, cytosolic (DYHC) (Cytoplasmic dynein heavy chain) (MAP 1C) Length = 4644 Score = 322 bits (825), Expect = 1e-87 Identities = 172/390 (44%), Positives = 243/390 (62%), Gaps = 5/390 (1%) Frame = +1 Query: 1 QRLLNSFLEYLFNEKCFSNNFALTFENDDKTEIVFPEFNNHGDILQWIYKLPDSQTPSWL 180 QRLLN+FLE LF + F + F L + D +I P+ + +QW+ LPD+QTPSWL Sbjct: 4260 QRLLNTFLERLFTTRSFDSEFKLACKVDGHKDIQMPDGIRREEFVQWVELLPDAQTPSWL 4319 Query: 181 GLPNSAEKVLLTNLGQSLIGNILKMQQTLEDDEEIVVDQRKSKRTIELGDSRPVWMRQLY 360 GLPN+AE+VLLT G +I +LKMQ ++D+ + K RT D RP WMR L+ Sbjct: 4320 GLPNNAERVLLTTQGVDMISKMLKMQMLEDEDDLAYAETEKKTRTDFTSDGRPAWMRTLH 4379 Query: 361 QSTQTWLQLLPKVVIPLQRTTENIKDPIYRCFEREVNSGSNLLLIVRKDLEDISEVCSGT 540 + WL L+P+ + PL+RT ENIKDP++R FEREV G+ LL VR+DL D+ +VC G Sbjct: 4380 TTASNWLHLIPQTLSPLKRTVENIKDPLFRFFEREVKMGAKLLQDVRQDLADVVQVCEGK 4439 Query: 541 KKPTNHHRSMMTNLAKGIIPISWIKYPISSGLTVIQWIHDFADRVKQLIHISNTAINSGA 720 KK TN+ R+++ L KGI+P SW Y + +G+TVIQW+ DF++R+KQL +IS A GA Sbjct: 4440 KKQTNYLRTLINELVKGILPRSWSHYTVPAGMTVIQWVSDFSERIKQLQNISQAAAAGGA 4499 Query: 721 AGLRSVKIWLGGLFMAEAYTTAIRQAVAQSNKWALEELFLDVELGEGK---IDKNTCIVT 891 L+++ + LG LF+ EAY TA RQ VAQ+N W+LEEL L+V + + +D + VT Sbjct: 4500 KELKNIHVCLGALFVPEAYITATRQYVAQANSWSLEELCLEVNVTASQSTTLDACSFGVT 4559 Query: 892 GLRLMGSEIVDSKTLKLSTTIYKDIDICRLKWIRIEKFEHFEENLIRDKVCLPVYLNAAR 1071 GL+L G+ ++K L LS I + + +L+W + E V LPVYLN R Sbjct: 4560 GLKLQGATCSNNK-LSLSNAISTVLPLTQLRWGKQTSAEK-----KASVVTLPVYLNFTR 4613 Query: 1072 TSILFTLYFN--TTDFINDFYKRGVAILAS 1155 ++FT+ F T + FY+RGVA+L + Sbjct: 4614 ADLIFTVDFEIATKEDPRSFYERGVAVLCT 4643
>sp|Q19020|DYHC_CAEEL Dynein heavy chain, cytosolic (DYHC) Length = 4568 Score = 255 bits (651), Expect = 2e-67 Identities = 144/386 (37%), Positives = 215/386 (55%) Frame = +1 Query: 1 QRLLNSFLEYLFNEKCFSNNFALTFENDDKTEIVFPEFNNHGDILQWIYKLPDSQTPSWL 180 Q LL+ LE LF K F + L + D + P + ++ W+ +L + Q P+WL Sbjct: 4210 QVLLDCVLENLFTAKSFEQDHVLIPKYDGDDSLFTPNMSKKDQMIGWVEELKNEQLPAWL 4269 Query: 181 GLPNSAEKVLLTNLGQSLIGNILKMQQTLEDDEEIVVDQRKSKRTIELGDSRPVWMRQLY 360 GLPN+AEKVLLT G+S++ N+LK+ DEE+ ++ + +P WM QL Sbjct: 4270 GLPNNAEKVLLTKRGESMLRNMLKVT-----DEELAFNEDGKEEV------KPQWMAQLG 4318 Query: 361 QSTQTWLQLLPKVVIPLQRTTENIKDPIYRCFEREVNSGSNLLLIVRKDLEDISEVCSGT 540 + + WLQLLPK ++ ++RT ENIKDP++R FEREVN GS LL +R+DL +IS VC Sbjct: 4319 ELAKQWLQLLPKEIVKMRRTVENIKDPLFRFFEREVNLGSQLLKDIRRDLNEISAVCRAE 4378 Query: 541 KKPTNHHRSMMTNLAKGIIPISWIKYPISSGLTVIQWIHDFADRVKQLIHISNTAINSGA 720 KK N R++ +L KG +P W +Y + +TV+ W+ D +R+KQLI I GA Sbjct: 4379 KKQNNETRALAASLQKGEVPTGWKRYTVpreVTVMDWMTDLNERLKQLIRI------GGA 4432 Query: 721 AGLRSVKIWLGGLFMAEAYTTAIRQAVAQSNKWALEELFLDVELGEGKIDKNTCIVTGLR 900 L+ WLGG F EAY TA RQ VAQ+N W+LE+L L + +G + ++G+ Sbjct: 4433 DNLKRETFWLGGTFSPEAYITATRQQVAQANTWSLEQLNLHIHIGRTD-STDVFRISGID 4491 Query: 901 LMGSEIVDSKTLKLSTTIYKDIDICRLKWIRIEKFEHFEENLIRDKVCLPVYLNAARTSI 1080 + G++ V L+L + + DI W + + D LP+YL R + Sbjct: 4492 IRGAKSVGGNKLELCELVKSECDIVEFSW----------KQDVADGTRLPLYLYGDRRQL 4541 Query: 1081 LFTLYFNTTDFINDFYKRGVAILASS 1158 + L F+ + FY+RGVA++A+S Sbjct: 4542 ISPLAFHLSS-ATVFYQRGVALVANS 4566
>sp|P34036|DYHC_DICDI Dynein heavy chain, cytosolic (DYHC) Length = 4725 Score = 219 bits (559), Expect = 1e-56 Identities = 139/386 (36%), Positives = 214/386 (55%), Gaps = 5/386 (1%) Frame = +1 Query: 4 RLLNSFLEYLFNEKCFSNNFALTFENDDKTEIVFPEFNNHGDILQWIYKLPDSQTPSWLG 183 RLL SFLE LF F+ +F L + PE ++WI LP+ TP WLG Sbjct: 4349 RLLYSFLEQLFTPSAFNPDFPLV----PSIGLSVPEGTTRAHFMKWIEALPEISTPIWLG 4404 Query: 184 LPNSAEKVLLTNLGQSLIGNILKMQQTLEDDEEIVVDQRKSKRTIELGDSRPVWMRQLYQ 363 LP +AE +LL+N + +I ++ KMQ + ED E+ V SK+ S +L Sbjct: 4405 LPENAESLLLSNKARKMINDLQKMQSSEEDGEDDQVSG-SSKKESSSSSSEDKGKAKLRA 4463 Query: 364 STQTWLQLLPKVVIPLQRTTENIKDPIYRCFEREVNSGSNLLLIVRKDLEDISEVCSGTK 543 + W +LLPK + L+RTT+NIKDP++RCFERE+++G L+ + DL ++ E+ SG Sbjct: 4464 TITEWTKLLPKPLKQLKRTTQNIKDPLFRCFEREISTGGKLVKKITNDLANLLELISGNI 4523 Query: 544 KPTNHHRSMMTNLAKGIIPISWIKYPISSGLTVIQWIHDFADRVKQLIHISNTAINSGAA 723 K TN+ RS+ T+++KGI+P W Y + +++ WI DF+ R++QL IS ++ S Sbjct: 4524 KSTNYLRSLTTSISKGIVPKEWKWYSVPETISLSVWISDFSKRMQQLSEISESSDYS--- 4580 Query: 724 GLRSVKIWLGGLFMAEAYTTAIRQAVAQSNKWALEELFLDVELGEGKIDKN---TCIVTG 894 S+++WLGGL EAY TA RQ+ +Q N W+LE L L GKI + V G Sbjct: 4581 ---SIQVWLGGLLNPEAYITATRQSASQLNGWSLENLRLHAS-SLGKISSEGGASFNVKG 4636 Query: 895 LRLMGSEIVDSKTLKLSTTIYKDIDICRLKWIRIEKFEHFEENLIRDKVCLPVYLNAART 1074 + L G+ + ++ L + + I I L W +K + N K+ +PVYLN R+ Sbjct: 4637 MALEGA-VWNNDQLTPTDILSTPISIATLTW--KDKDDPIFNN-SSSKLSVPVYLNETRS 4692 Query: 1075 SILFT--LYFNTTDFINDFYKRGVAI 1146 +LF+ L ++ + ++Y+R V+I Sbjct: 4693 ELLFSIDLPYDQSTSKQNWYQRSVSI 4718
>sp|Q27171|DYHC_PARTE Dynein heavy chain, cytosolic (DYHC) (DHC08) Length = 4540 Score = 208 bits (529), Expect = 3e-53 Identities = 118/366 (32%), Positives = 207/366 (56%), Gaps = 2/366 (0%) Frame = +1 Query: 1 QRLLNSFLEYLFNEKCFSNNFALTFENDDKTEIVFPEFNNHGDILQWIYKLPDSQTPSWL 180 Q++L S +E F E+ F++N L F + K I PE + D +QWI +LP +++P W Sbjct: 4163 QKILQSLVEQFFTEQSFNHNHPLFFTLEGKEAITVPEGRTYLDFMQWIEQLPKTESPEWS 4222 Query: 181 GLPNSAEKVLLTNLGQSLIGNILKMQQTLEDD-EEIVVDQRKSKRTIELGDSRPVWMRQL 357 GLP++ E+V L Q LI + +QQ E++ +I V K+++ + W++ L Sbjct: 4223 GLPSNVERVQRDQLTQKLITKVQNLQQEGEEEITQIEVQTEKTQKKDNKKSDQVQWLQDL 4282 Query: 358 YQSTQTWLQLLPKVVIPLQRTTENIKDPIYRCFEREVNSGSNLLLIVRKDLEDISEVCSG 537 + + + +LP + PL+RT ++I DP++R +RE+ S LL VR+++E++ ++ G Sbjct: 4283 LEKVEKFKAILPNKISPLERTADSINDPLFRFLDREITVASKLLKAVRQNIEELIQLAQG 4342 Query: 538 TKKPTNHHRSMMTNLAKGIIPISWIKYPISSGLTVIQWIHDFADRVKQLIHISNTAINSG 717 TN R + ++ I+P W KY + + + + W+ DF R+ Q + T Sbjct: 4343 KILATNILRQLAKDVFNNIVPAQWNKYNVIT-MPLNDWVGDFKRRIDQFDLLGKT----- 4396 Query: 718 AAGLRSVKIWLGGLFMAEAYTTAIRQAVAQSNKWALEELFLDVELGEGKIDKNTCIVTGL 897 + ++W GGL EAY TA RQ VAQ+NKW+LEEL L + + ID+++ ++ G+ Sbjct: 4397 -KDFQKGQVWFGGLLFPEAYLTATRQYVAQANKWSLEELELQMIPEDQGIDEDSFVIEGV 4455 Query: 898 RLMGSEIVDSKTLKLSTTIYKDIDICRLKWIRIEKFEHFEENLI-RDKVCLPVYLNAART 1074 + G + DSKTL++ I +I + LK I ++ + ++ ++ D++ LPVYLN R Sbjct: 4456 SMEGGHL-DSKTLQV--RIVNEISVA-LKPITLKWCKTSQKGVVGDDEIVLPVYLNKTRK 4511 Query: 1075 SILFTL 1092 +++F+L Sbjct: 4512 NLIFSL 4517
>sp|Q8VHE6|DYH5_MOUSE Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (Mdnah5) Length = 4621 Score = 65.5 bits (158), Expect = 3e-10 Identities = 93/402 (23%), Positives = 157/402 (39%), Gaps = 19/402 (4%) Frame = +1 Query: 1 QRLLNSFLEYLFNEKCFSNNFALTFENDDKTEIVFPEFNNHGDILQWIYKLPDSQTPSWL 180 +RLLN+F + F+E F +F TF P+ + LQ+I LP +P Sbjct: 4258 KRLLNTFAKVWFSENMFGPDF--TFYQGYN----IPKCSTVDGYLQYIQSLPAYDSPEVF 4311 Query: 181 GLPNSAEKVLLTNLGQSLIGNILKMQQTLEDDEEIVVDQRKSKRTIELGDSRPVWMRQLY 360 GL +A+ + L + ++ IL +Q D D+ + L D Sbjct: 4312 GLHPNADITYQSKLAKDVLDTILGIQPK---DSSGGGDETREAVVARLADDM-------- 4360 Query: 361 QSTQTWLQLLPKVVIPLQRTTENIK----DPIYRCFEREVNSGSNLLLIVRKDLEDISEV 528 L+ LP+ P + K P+ +E++ +L +VR L ++ Sbjct: 4361 ------LEKLPEDYSPFEVKERLQKMGPFQPMNIFLRQEIDRMQRVLSLVRSTLTELKLA 4414 Query: 529 CSGTKKPTNHHRSMMTNLAKGIIPISWIKYPISSGLTVIQWIHDFADRVKQLIHISNTAI 708 GT + + R + + IP W K S T+ W + +R N Sbjct: 4415 VDGTIIMSENLRDALDCMFDARIPARWKKASWVSS-TLGFWFTELLER--------NCQF 4465 Query: 709 NSGAAGLRSVKIWLGGLFMAEAYTTAIRQAVAQSNK-WALEELFLDVELGEGKIDKNTC- 882 S + R W+ G F + + TA+RQ + ++NK WAL+ + L E+ + D + Sbjct: 4466 TSWVSNGRPHCFWMTGFFNPQGFLTAMRQEITRANKGWALDNMVLCNEVTKFMKDDISAP 4525 Query: 883 -----IVTGLRLMGSEIVDSKTLKL----STTIYKDIDICRLKWIRIEKFEHFEENLIRD 1035 V GL L G+ D + +KL +++ + + R+ E N RD Sbjct: 4526 PTEGVYVYGLYLEGAG-WDKRNMKLIESKPKVLFELMPVIRI---------FAENNTARD 4575 Query: 1036 K--VCLPVYLNAARTSI--LFTLYFNTTDFINDFYKRGVAIL 1149 C P+Y RT + + + T + RGVA+L Sbjct: 4576 PRLYCCPIYKKPVRTDLNYIAAVDLKTAQAPEHWVLRGVALL 4617
>sp|Q8TE73|DYH5_HUMAN Ciliary dynein heavy chain 5 (Axonemal beta dynein heavy chain 5) (HL1) Length = 4624 Score = 64.3 bits (155), Expect = 7e-10 Identities = 90/402 (22%), Positives = 159/402 (39%), Gaps = 19/402 (4%) Frame = +1 Query: 1 QRLLNSFLEYLFNEKCFSNNFALTFENDDKTEIVFPEFNNHGDILQWIYKLPDSQTPSWL 180 +RLLN+F + F+E F +F+ + P+ + + LQ+I LP +P Sbjct: 4261 KRLLNTFAKVWFSENMFGPDFSFYQGYN------IPKCSTVDNYLQYIQSLPAYDSPEVF 4314 Query: 181 GLPNSAEKVLLTNLGQSLIGNILKMQQTLEDDEEIVVDQRKSKRTIELGDSRPVWMRQLY 360 GL +A+ + L + ++ IL +Q D D+ + L D Sbjct: 4315 GLHPNADITYQSKLAKDVLDTILGIQPK---DTSGGGDETREAVVARLADDM-------- 4363 Query: 361 QSTQTWLQLLPKVVIPLQRTTENIK----DPIYRCFEREVNSGSNLLLIVRKDLEDISEV 528 L+ LP +P + K P+ +E++ +L +VR L ++ Sbjct: 4364 ------LEKLPPDYVPFEVKERLQKMGPFQPMNIFLRQEIDRMQRVLSLVRSTLTELKLA 4417 Query: 529 CSGTKKPTNHHRSMMTNLAKGIIPISWIKYPISSGLTVIQWIHDFADRVKQLIHISNTAI 708 GT + + R + + IP W K S T+ W + +R N+ Sbjct: 4418 IDGTIIMSENLRDALDCMFDARIPAWWKKASWISS-TLGFWFTELIER--------NSQF 4468 Query: 709 NSGAAGLRSVKIWLGGLFMAEAYTTAIRQAVAQSNK-WALEELFLDVELGEGKIDKNTC- 882 S R W+ G F + + TA+RQ + ++NK WAL+ + L E+ + D + Sbjct: 4469 TSWVFNGRPHCFWMTGFFNPQGFLTAMRQEITRANKGWALDNMVLCNEVTKWMKDDISAP 4528 Query: 883 -----IVTGLRLMGSEIVDSKTLKL----STTIYKDIDICRLKWIRIEKFEHFEENLIRD 1035 V GL L G+ D + +KL +++ + + R+ + E N +RD Sbjct: 4529 PTEGVYVYGLYLEGAG-WDKRNMKLIESKPKVLFELMPVIRI---------YAENNTLRD 4578 Query: 1036 K--VCLPVYLNAARTSI--LFTLYFNTTDFINDFYKRGVAIL 1149 P+Y RT + + + T + RGVA+L Sbjct: 4579 PRFYSCPIYKKPVRTDLNYIAAVDLRTAQTPEHWVLRGVALL 4620
>sp|P78716|DYHC_FUSSO Dynein heavy chain, cytosolic (DYHC) Length = 4349 Score = 64.3 bits (155), Expect = 7e-10 Identities = 28/66 (42%), Positives = 47/66 (71%) Frame = +1 Query: 97 IVFPEFNNHGDILQWIYKLPDSQTPSWLGLPNSAEKVLLTNLGQSLIGNILKMQQTLEDD 276 +V P + D WI++LP+ + P++LGLP +AEK+LL LG+SLIGN+ K+ + L++ Sbjct: 4283 LVVPSGTSFQDFNAWIHRLPEREPPTYLGLPANAEKLLLGGLGRSLIGNLRKVTELLDEG 4342 Query: 277 EEIVVD 294 E++V + Sbjct: 4343 EQLVTE 4348
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 133,733,503
Number of Sequences: 369166
Number of extensions: 2661067
Number of successful extensions: 6817
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 6493
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6785
length of database: 68,354,980
effective HSP length: 113
effective length of database: 47,479,925
effective search space used: 14576336975
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail