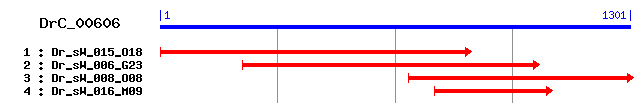
DrC_00606
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00606 (1301 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P52193|CRTC1_BOVIN Calreticulin, brain isoform 1 precurs... 376 e-104 sp|P42918|CRTC2_BOVIN Calreticulin, brain isoform 2 precurs... 375 e-103 sp|P27797|CRTC_HUMAN Calreticulin precursor (CRP55) (Calreg... 375 e-103 sp|P14211|CRTC_MOUSE Calreticulin precursor (CRP55) (Calreg... 372 e-102 sp|Q8K3H7|CRTC_CRIGR Calreticulin precursor (CRP55) (Calreg... 370 e-102 sp|P18418|CRTC_RAT Calreticulin precursor (CRP55) (Calregul... 370 e-102 sp|P15253|CRTC_RABIT Calreticulin precursor (CRP55) (Calreg... 369 e-102 sp|P11012|RAL1_ONCVO RAL-1 protein precursor (RAL1 antigen)... 365 e-100 sp|P29413|CRTC_DROME Calreticulin precursor (CRP55) (Calreg... 364 e-100 sp|Q06814|CRTC_SCHMA Calreticulin precursor (SM4 protein) 341 2e-93
>sp|P52193|CRTC1_BOVIN Calreticulin, brain isoform 1 precursor (CRP55) (Calregulin) (HACBP) Length = 417 Score = 376 bits (966), Expect = e-104 Identities = 170/251 (67%), Positives = 199/251 (79%) Frame = +1 Query: 268 LMSSDIDQTKFHGETPYHIMFGPDICGLSTKKVHVIFNYKGQNHLIKKEIRCKDDEFSHV 447 L + +DQT HG++ Y+IMFGPDICG TKKVHVIFNYKG+N LI K+IRCKDDEF+H+ Sbjct: 112 LFPAGLDQTDMHGDSEYNIMFGPDICGPGTKKVHVIFNYKGKNVLINKDIRCKDDEFTHL 171 Query: 448 YTLIVSPDNTYEVLIDNEKVESGNLEDDWDFLAPKKIKDPNAKKPEDWEDNEKIDDSTDK 627 YTLIV P+NTYEV IDN +VESG+LEDDWDFL PKKIKDP+A KPEDW+D KIDD TD Sbjct: 172 YTLIVRPNNTYEVKIDNSQVESGSLEDDWDFLPPKKIKDPDAAKPEDWDDRAKIDDPTDS 231 Query: 628 KPEDWDKPKTIADPKAKKPXXXXXXXXXXXXAPQIDNPEYKGEWKPKQIDNPKYKGPWIH 807 KPEDWDKP+ I DP AKKP P I NPEYKGEWKP+QIDNP+YKG WIH Sbjct: 232 KPEDWDKPEHIPDPDAKKPEDWDEEMDGEWEPPVIQNPEYKGEWKPRQIDNPEYKGIWIH 291 Query: 808 PEIDNPDYKPESNLYLYKNIGSIGFDLWQVKSGTIFDNILLTDDPAYAKKYVEKTWKETA 987 PEIDNP+Y P+SN+Y Y+N +G DLWQVKSGTIFDN L+T+D AYA+++ +TW T Sbjct: 292 PEIDNPEYSPDSNIYAYENFAVLGLDLWQVKSGTIFDNFLITNDEAYAEEFGNETWGVTK 351 Query: 988 AGEKKMKDKQD 1020 A EK+MKDKQD Sbjct: 352 AAEKQMKDKQD 362
Score = 109 bits (273), Expect = 2e-23
Identities = 53/90 (58%), Positives = 69/90 (76%), Gaps = 1/90 (1%)
Frame = +3
Query: 3 EEFLT-DSYKTQWVESTYKGADQGKFTWTAGKFYNDAEKDKGIQTSQDAKFYGISTKFDK 179
E+FL D + +W+ES +K D GKF ++GKFY D EKDKG+QTSQDA+FY +S +F+
Sbjct: 25 EQFLDGDGWTERWIESKHK-PDFGKFVLSSGKFYGDQEKDKGLQTSQDARFYALSARFE- 82
Query: 180 QFSNEGKKLVIQFQVKHEQTIDCGGGYVNI 269
FSN+G+ LV+QF VKHEQ IDCGGGYV +
Sbjct: 83 PFSNKGQTLVVQFTVKHEQNIDCGGGYVKL 112
>sp|P42918|CRTC2_BOVIN Calreticulin, brain isoform 2 precursor (CRP55) (Calregulin) (HACBP) Length = 421 Score = 375 bits (963), Expect = e-103 Identities = 169/246 (68%), Positives = 197/246 (80%) Frame = +1 Query: 283 IDQTKFHGETPYHIMFGPDICGLSTKKVHVIFNYKGQNHLIKKEIRCKDDEFSHVYTLIV 462 +DQT HG++ Y+IMFGPDICG TKKVHVIFNYKG+N LI K+IRCKDDEF+H+YTLIV Sbjct: 121 LDQTDMHGDSEYNIMFGPDICGPGTKKVHVIFNYKGKNVLINKDIRCKDDEFTHLYTLIV 180 Query: 463 SPDNTYEVLIDNEKVESGNLEDDWDFLAPKKIKDPNAKKPEDWEDNEKIDDSTDKKPEDW 642 P+NTYEV IDN +VESG+LEDDWDFL PKKIKDP+A KPEDW+D KIDD TD KPEDW Sbjct: 181 RPNNTYEVKIDNSQVESGSLEDDWDFLPPKKIKDPDAAKPEDWDDRAKIDDPTDSKPEDW 240 Query: 643 DKPKTIADPKAKKPXXXXXXXXXXXXAPQIDNPEYKGEWKPKQIDNPKYKGPWIHPEIDN 822 DKP+ I DP AKKP P I NPEYKGEWKP+QIDNP+YKG WIHPEIDN Sbjct: 241 DKPEHIPDPDAKKPEDWDEEMDGEWEPPLIQNPEYKGEWKPRQIDNPEYKGIWIHPEIDN 300 Query: 823 PDYKPESNLYLYKNIGSIGFDLWQVKSGTIFDNILLTDDPAYAKKYVEKTWKETAAGEKK 1002 P+Y P+SN+Y Y+N +G DLWQVKSGTIFDN L+T+D AYA+++ +TW T A EK+ Sbjct: 301 PEYSPDSNIYAYENFAVLGLDLWQVKSGTIFDNFLITNDEAYAEEFGNETWGVTKAAEKQ 360 Query: 1003 MKDKQD 1020 MKDKQD Sbjct: 361 MKDKQD 366
>sp|P27797|CRTC_HUMAN Calreticulin precursor (CRP55) (Calregulin) (HACBP) (ERp60) (grp60) Length = 417 Score = 375 bits (963), Expect = e-103 Identities = 169/251 (67%), Positives = 197/251 (78%) Frame = +1 Query: 268 LMSSDIDQTKFHGETPYHIMFGPDICGLSTKKVHVIFNYKGQNHLIKKEIRCKDDEFSHV 447 L + +DQT HG++ Y+IMFGPDICG TKKVHVIFNYKG+N LI K+IRCKDDEF+H+ Sbjct: 112 LFPNSLDQTDMHGDSEYNIMFGPDICGPGTKKVHVIFNYKGKNVLINKDIRCKDDEFTHL 171 Query: 448 YTLIVSPDNTYEVLIDNEKVESGNLEDDWDFLAPKKIKDPNAKKPEDWEDNEKIDDSTDK 627 YTLIV PDNTYEV IDN +VESG+LEDDWDFL PKKIKDP+A KPEDW++ KIDD TD Sbjct: 172 YTLIVRPDNTYEVKIDNSQVESGSLEDDWDFLPPKKIKDPDASKPEDWDERAKIDDPTDS 231 Query: 628 KPEDWDKPKTIADPKAKKPXXXXXXXXXXXXAPQIDNPEYKGEWKPKQIDNPKYKGPWIH 807 KPEDWDKP+ I DP AKKP P I NPEYKGEWKP+QIDNP YKG WIH Sbjct: 232 KPEDWDKPEHIPDPDAKKPEDWDEEMDGEWEPPVIQNPEYKGEWKPRQIDNPDYKGTWIH 291 Query: 808 PEIDNPDYKPESNLYLYKNIGSIGFDLWQVKSGTIFDNILLTDDPAYAKKYVEKTWKETA 987 PEIDNP+Y P+ ++Y Y N G +G DLWQVKSGTIFDN L+T+D AYA+++ +TW T Sbjct: 292 PEIDNPEYSPDPSIYAYDNFGVLGLDLWQVKSGTIFDNFLITNDEAYAEEFGNETWGVTK 351 Query: 988 AGEKKMKDKQD 1020 A EK+MKDKQD Sbjct: 352 AAEKQMKDKQD 362
Score = 110 bits (275), Expect = 9e-24
Identities = 53/90 (58%), Positives = 70/90 (77%), Gaps = 1/90 (1%)
Frame = +3
Query: 3 EEFLT-DSYKTQWVESTYKGADQGKFTWTAGKFYNDAEKDKGIQTSQDAKFYGISTKFDK 179
E+FL D + ++W+ES +K +D GKF ++GKFY D EKDKG+QTSQDA+FY +S F+
Sbjct: 25 EQFLDGDGWTSRWIESKHK-SDFGKFVLSSGKFYGDEEKDKGLQTSQDARFYALSASFE- 82
Query: 180 QFSNEGKKLVIQFQVKHEQTIDCGGGYVNI 269
FSN+G+ LV+QF VKHEQ IDCGGGYV +
Sbjct: 83 PFSNKGQTLVVQFTVKHEQNIDCGGGYVKL 112
>sp|P14211|CRTC_MOUSE Calreticulin precursor (CRP55) (Calregulin) (HACBP) (ERp60) Length = 416 Score = 372 bits (954), Expect = e-102 Identities = 168/251 (66%), Positives = 196/251 (78%) Frame = +1 Query: 268 LMSSDIDQTKFHGETPYHIMFGPDICGLSTKKVHVIFNYKGQNHLIKKEIRCKDDEFSHV 447 L S +DQ HG++ Y+IMFGPDICG TKKVHVIFNYKG+N LI K+IRCKDDEF+H+ Sbjct: 112 LFPSGLDQKDMHGDSEYNIMFGPDICGPGTKKVHVIFNYKGKNVLINKDIRCKDDEFTHL 171 Query: 448 YTLIVSPDNTYEVLIDNEKVESGNLEDDWDFLAPKKIKDPNAKKPEDWEDNEKIDDSTDK 627 YTLIV PDNTYEV IDN +VESG+LEDDWDFL PKKIKDP+A KPEDW++ KIDD TD Sbjct: 172 YTLIVRPDNTYEVKIDNSQVESGSLEDDWDFLPPKKIKDPDAAKPEDWDERAKIDDPTDS 231 Query: 628 KPEDWDKPKTIADPKAKKPXXXXXXXXXXXXAPQIDNPEYKGEWKPKQIDNPKYKGPWIH 807 KPEDWDKP+ I DP AKKP P I NPEYKGEWKP+QIDNP YKG WIH Sbjct: 232 KPEDWDKPEHIPDPDAKKPEDWDEEMDGEWEPPVIQNPEYKGEWKPRQIDNPDYKGTWIH 291 Query: 808 PEIDNPDYKPESNLYLYKNIGSIGFDLWQVKSGTIFDNILLTDDPAYAKKYVEKTWKETA 987 PEIDNP+Y P++N+Y Y + +G DLWQVKSGTIFDN L+T+D AYA+++ +TW T Sbjct: 292 PEIDNPEYSPDANIYAYDSFAVLGLDLWQVKSGTIFDNFLITNDEAYAEEFGNETWGVTK 351 Query: 988 AGEKKMKDKQD 1020 A EK+MKDKQD Sbjct: 352 AAEKQMKDKQD 362
Score = 112 bits (281), Expect = 2e-24
Identities = 55/90 (61%), Positives = 71/90 (78%), Gaps = 1/90 (1%)
Frame = +3
Query: 3 EEFLT-DSYKTQWVESTYKGADQGKFTWTAGKFYNDAEKDKGIQTSQDAKFYGISTKFDK 179
E+FL D++ +WVES +K +D GKF ++GKFY D EKDKG+QTSQDA+FY +S KF+
Sbjct: 25 EQFLDGDAWTNRWVESKHK-SDFGKFVLSSGKFYGDLEKDKGLQTSQDARFYALSAKFE- 82
Query: 180 QFSNEGKKLVIQFQVKHEQTIDCGGGYVNI 269
FSN+G+ LV+QF VKHEQ IDCGGGYV +
Sbjct: 83 PFSNKGQTLVVQFTVKHEQNIDCGGGYVKL 112
>sp|Q8K3H7|CRTC_CRIGR Calreticulin precursor (CRP55) (Calregulin) (HACBP) (ERp60) Length = 417 Score = 370 bits (951), Expect = e-102 Identities = 167/251 (66%), Positives = 195/251 (77%) Frame = +1 Query: 268 LMSSDIDQTKFHGETPYHIMFGPDICGLSTKKVHVIFNYKGQNHLIKKEIRCKDDEFSHV 447 L +DQ HG++ Y+IMFGPDICG TKKVHVIFNYKG+N LI K+IRCKDDEF+H+ Sbjct: 112 LFPGSLDQKDMHGDSEYNIMFGPDICGPGTKKVHVIFNYKGKNVLINKDIRCKDDEFTHL 171 Query: 448 YTLIVSPDNTYEVLIDNEKVESGNLEDDWDFLAPKKIKDPNAKKPEDWEDNEKIDDSTDK 627 YTLIV PDNTYEV IDN +VESG+LEDDWDFL PKKIKDP+A KPEDW++ KIDD TD Sbjct: 172 YTLIVRPDNTYEVKIDNSQVESGSLEDDWDFLPPKKIKDPDAAKPEDWDERAKIDDPTDS 231 Query: 628 KPEDWDKPKTIADPKAKKPXXXXXXXXXXXXAPQIDNPEYKGEWKPKQIDNPKYKGPWIH 807 KPEDWDKP+ I DP AKKP P I NPEYKGEWKP+QIDNP YKG WIH Sbjct: 232 KPEDWDKPEHIPDPDAKKPEDWDEEMDGEWEPPVIQNPEYKGEWKPRQIDNPDYKGTWIH 291 Query: 808 PEIDNPDYKPESNLYLYKNIGSIGFDLWQVKSGTIFDNILLTDDPAYAKKYVEKTWKETA 987 PEIDNP+Y P++N+Y Y + +G DLWQVKSGTIFDN L+T+D AYA+++ +TW T Sbjct: 292 PEIDNPEYSPDANIYAYDSFAVLGLDLWQVKSGTIFDNFLITNDEAYAEEFGNETWGVTK 351 Query: 988 AGEKKMKDKQD 1020 A EK+MKDKQD Sbjct: 352 ASEKQMKDKQD 362
Score = 111 bits (277), Expect = 5e-24
Identities = 54/90 (60%), Positives = 70/90 (77%), Gaps = 1/90 (1%)
Frame = +3
Query: 3 EEFLT-DSYKTQWVESTYKGADQGKFTWTAGKFYNDAEKDKGIQTSQDAKFYGISTKFDK 179
E+FL D + +WVES +K +D GKF ++GKFY D EKDKG+QTSQDA+FY +S +F+
Sbjct: 25 EQFLDGDDWTNRWVESKHK-SDFGKFVLSSGKFYGDQEKDKGLQTSQDARFYALSARFE- 82
Query: 180 QFSNEGKKLVIQFQVKHEQTIDCGGGYVNI 269
FSN+G+ LV+QF VKHEQ IDCGGGYV +
Sbjct: 83 PFSNKGQTLVVQFTVKHEQNIDCGGGYVKL 112
>sp|P18418|CRTC_RAT Calreticulin precursor (CRP55) (Calregulin) (HACBP) (ERp60) (CALBP) (Calcium-binding protein 3) (CABP3) Length = 416 Score = 370 bits (950), Expect = e-102 Identities = 167/251 (66%), Positives = 195/251 (77%) Frame = +1 Query: 268 LMSSDIDQTKFHGETPYHIMFGPDICGLSTKKVHVIFNYKGQNHLIKKEIRCKDDEFSHV 447 L +DQ HG++ Y+IMFGPDICG TKKVHVIFNYKG+N LI K+IRCKDDEF+H+ Sbjct: 112 LFPGGLDQKDMHGDSEYNIMFGPDICGPGTKKVHVIFNYKGKNVLINKDIRCKDDEFTHL 171 Query: 448 YTLIVSPDNTYEVLIDNEKVESGNLEDDWDFLAPKKIKDPNAKKPEDWEDNEKIDDSTDK 627 YTLIV PDNTYEV IDN +VESG+LEDDWDFL PKKIKDP+A KPEDW++ KIDD TD Sbjct: 172 YTLIVRPDNTYEVKIDNSQVESGSLEDDWDFLPPKKIKDPDAAKPEDWDERAKIDDPTDS 231 Query: 628 KPEDWDKPKTIADPKAKKPXXXXXXXXXXXXAPQIDNPEYKGEWKPKQIDNPKYKGPWIH 807 KPEDWDKP+ I DP AKKP P I NPEYKGEWKP+QIDNP YKG WIH Sbjct: 232 KPEDWDKPEHIPDPDAKKPEDWDEEMDGEWEPPVIQNPEYKGEWKPRQIDNPDYKGTWIH 291 Query: 808 PEIDNPDYKPESNLYLYKNIGSIGFDLWQVKSGTIFDNILLTDDPAYAKKYVEKTWKETA 987 PEIDNP+Y P++N+Y Y + +G DLWQVKSGTIFDN L+T+D AYA+++ +TW T Sbjct: 292 PEIDNPEYSPDANIYAYDSFAVLGLDLWQVKSGTIFDNFLITNDEAYAEEFGNETWGVTK 351 Query: 988 AGEKKMKDKQD 1020 A EK+MKDKQD Sbjct: 352 AAEKQMKDKQD 362
Score = 111 bits (278), Expect = 4e-24
Identities = 54/90 (60%), Positives = 71/90 (78%), Gaps = 1/90 (1%)
Frame = +3
Query: 3 EEFLT-DSYKTQWVESTYKGADQGKFTWTAGKFYNDAEKDKGIQTSQDAKFYGISTKFDK 179
E+FL D++ +WVES +K +D GKF ++GKFY D EKDKG+QTSQDA+FY +S +F+
Sbjct: 25 EQFLDGDAWTNRWVESKHK-SDFGKFVLSSGKFYGDQEKDKGLQTSQDARFYALSARFE- 82
Query: 180 QFSNEGKKLVIQFQVKHEQTIDCGGGYVNI 269
FSN+G+ LV+QF VKHEQ IDCGGGYV +
Sbjct: 83 PFSNKGQTLVVQFTVKHEQNIDCGGGYVKL 112
>sp|P15253|CRTC_RABIT Calreticulin precursor (CRP55) (Calregulin) (HACBP) (ERp60) Length = 418 Score = 369 bits (948), Expect = e-102 Identities = 166/251 (66%), Positives = 195/251 (77%) Frame = +1 Query: 268 LMSSDIDQTKFHGETPYHIMFGPDICGLSTKKVHVIFNYKGQNHLIKKEIRCKDDEFSHV 447 L + +DQ HG++ Y+IMFGPDICG TKKVHVIFNYKG+N LI K+IRCKDDEF+H+ Sbjct: 112 LFPAGLDQKDMHGDSEYNIMFGPDICGPGTKKVHVIFNYKGKNVLINKDIRCKDDEFTHL 171 Query: 448 YTLIVSPDNTYEVLIDNEKVESGNLEDDWDFLAPKKIKDPNAKKPEDWEDNEKIDDSTDK 627 YTLIV PDNTYEV IDN +VESG+LEDDWDFL PKKIKDP+A KPEDW++ KIDD TD Sbjct: 172 YTLIVRPDNTYEVKIDNSQVESGSLEDDWDFLPPKKIKDPDASKPEDWDERAKIDDPTDS 231 Query: 628 KPEDWDKPKTIADPKAKKPXXXXXXXXXXXXAPQIDNPEYKGEWKPKQIDNPKYKGPWIH 807 KPEDWDKP+ I DP AKKP P I NPEYKGEWKP+QIDNP YKG WIH Sbjct: 232 KPEDWDKPEHIPDPDAKKPEDWDEEMDGEWEPPVIQNPEYKGEWKPRQIDNPDYKGTWIH 291 Query: 808 PEIDNPDYKPESNLYLYKNIGSIGFDLWQVKSGTIFDNILLTDDPAYAKKYVEKTWKETA 987 PEIDNP+Y P++N+Y Y + +G DLWQVKSGTIFDN L+T+D AYA+++ +TW T Sbjct: 292 PEIDNPEYSPDANIYAYDSFAVLGLDLWQVKSGTIFDNFLITNDEAYAEEFGNETWGVTK 351 Query: 988 AGEKKMKDKQD 1020 EK+MKDKQD Sbjct: 352 TAEKQMKDKQD 362
Score = 110 bits (275), Expect = 9e-24
Identities = 53/90 (58%), Positives = 70/90 (77%), Gaps = 1/90 (1%)
Frame = +3
Query: 3 EEFLT-DSYKTQWVESTYKGADQGKFTWTAGKFYNDAEKDKGIQTSQDAKFYGISTKFDK 179
E+FL D + +W+ES +K +D GKF ++GKFY D EKDKG+QTSQDA+FY +S +F+
Sbjct: 25 EQFLDGDGWTERWIESKHK-SDFGKFVLSSGKFYGDQEKDKGLQTSQDARFYALSARFE- 82
Query: 180 QFSNEGKKLVIQFQVKHEQTIDCGGGYVNI 269
FSN+G+ LV+QF VKHEQ IDCGGGYV +
Sbjct: 83 PFSNKGQPLVVQFTVKHEQNIDCGGGYVKL 112
>sp|P11012|RAL1_ONCVO RAL-1 protein precursor (RAL1 antigen) (41 kDa larval antigen) Length = 388 Score = 365 bits (937), Expect = e-100 Identities = 183/316 (57%), Positives = 218/316 (68%), Gaps = 1/316 (0%) Frame = +1 Query: 70 ENSHGQRVNSIMTLKKIKESRLAKMLSSMA-FQQSLINNSAMKERSWLFNSK*NMNKQLI 246 E SHG+ + K +K ++ AK S A F +S N K +S + + + Sbjct: 47 EISHGKFYGDAVKDKGLKTTQDAKFYSIGAKFDKSFSN----KGKSLVIQFSVKHEQDID 102 Query: 247 VVVDMSILMSSDIDQTKFHGETPYHIMFGPDICGLSTKKVHVIFNYKGQNHLIKKEIRCK 426 LM+SD++ HGETPYHIMFGPDICG TKKVHVIF+YK +NH+IKK+IRCK Sbjct: 103 CGGGYVKLMASDVNLEDSHGETPYHIMFGPDICGPGTKKVHVIFHYKDRNHMIKKDIRCK 162 Query: 427 DDEFSHVYTLIVSPDNTYEVLIDNEKVESGNLEDDWDFLAPKKIKDPNAKKPEDWEDNEK 606 DD F+H+YTLIV+ DNTYEV ID EK ESG LE DWDFL PKKIKDP+AKKPEDW++ E Sbjct: 163 DDVFTHLYTLIVNSDNTYEVQIDGEKAESGELEADWDFLPPKKIKDPDAKKPEDWDEREF 222 Query: 607 IDDSTDKKPEDWDKPKTIADPKAKKPXXXXXXXXXXXXAPQIDNPEYKGEWKPKQIDNPK 786 IDD DKKPEDWDKP+ I DP AKKP P +DNPEYKGEWKPKQ NP Sbjct: 223 IDDEDDKKPEDWDKPEHIPDPDAKKPEDWDDEMDGEWEPPMVDNPEYKGEWKPKQKKNPA 282 Query: 787 YKGPWIHPEIDNPDYKPESNLYLYKNIGSIGFDLWQVKSGTIFDNILLTDDPAYAKKYVE 966 YKG WIHPEI+ PDY P+ NLY+Y +IG+IGFDLWQVKSGTIFD++++TD AKK+ E Sbjct: 283 YKGKWIHPEIEIPDYTPDDNLYVYDDIGAIGFDLWQVKSGTIFDDVIVTDSVEEAKKFGE 342 Query: 967 KTWKETAAGEKKMKDK 1014 KT K T GEKK K Sbjct: 343 KTLKITREGEKKKGKK 358
Score = 116 bits (290), Expect = 2e-25
Identities = 54/89 (60%), Positives = 68/89 (76%)
Frame = +3
Query: 3 EEFLTDSYKTQWVESTYKGADQGKFTWTAGKFYNDAEKDKGIQTSQDAKFYGISTKFDKQ 182
E+F D ++ +W++S +K D GK+ + GKFY DA KDKG++T+QDAKFY I KFDK
Sbjct: 23 EDFSDDDWEKRWIKSKHKD-DFGKWEISHGKFYGDAVKDKGLKTTQDAKFYSIGAKFDKS 81
Query: 183 FSNEGKKLVIQFQVKHEQTIDCGGGYVNI 269
FSN+GK LVIQF VKHEQ IDCGGGYV +
Sbjct: 82 FSNKGKSLVIQFSVKHEQDIDCGGGYVKL 110
>sp|P29413|CRTC_DROME Calreticulin precursor (CRP55) (Calregulin) (HACBP) Length = 406 Score = 364 bits (934), Expect = e-100 Identities = 170/251 (67%), Positives = 190/251 (75%) Frame = +1 Query: 268 LMSSDIDQTKFHGETPYHIMFGPDICGLSTKKVHVIFNYKGQNHLIKKEIRCKDDEFSHV 447 L +DQT HGE+PY IMFGPDICG TKKVHVIF+YKG+NHLI K+IRCKDD ++H Sbjct: 112 LFDCSLDQTDMHGESPYEIMFGPDICGPGTKKVHVIFSYKGKNHLISKDIRCKDDVYTHF 171 Query: 448 YTLIVSPDNTYEVLIDNEKVESGNLEDDWDFLAPKKIKDPNAKKPEDWEDNEKIDDSTDK 627 YTLIV PDNTYEVLIDNEKVESGNLEDDWDFLAPKKIKDP A KPEDW+D I D DK Sbjct: 172 YTLIVRPDNTYEVLIDNEKVESGNLEDDWDFLAPKKIKDPTATKPEDWDDRATIPDPDDK 231 Query: 628 KPEDWDKPKTIADPKAKKPXXXXXXXXXXXXAPQIDNPEYKGEWKPKQIDNPKYKGPWIH 807 KPEDWDKP+ I DP A KP P IDNPE+KGEW+PKQ+DNP YKG W H Sbjct: 232 KPEDWDKPEHIPDPDATKPEDWDDEMDGEWEPPMIDNPEFKGEWQPKQLDNPNYKGAWEH 291 Query: 808 PEIDNPDYKPESNLYLYKNIGSIGFDLWQVKSGTIFDNILLTDDPAYAKKYVEKTWKETA 987 PEI NP+Y P+ LYL K I ++GFDLWQVKSGTIFDN+L+TDD A K + K T Sbjct: 292 PEIANPEYVPDDKLYLRKEICTLGFDLWQVKSGTIFDNVLITDDVELAAKAAAEV-KNTQ 350 Query: 988 AGEKKMKDKQD 1020 AGEKKMK+ QD Sbjct: 351 AGEKKMKEAQD 361
Score = 112 bits (281), Expect = 2e-24
Identities = 54/89 (60%), Positives = 64/89 (71%)
Frame = +3
Query: 3 EEFLTDSYKTQWVESTYKGADQGKFTWTAGKFYNDAEKDKGIQTSQDAKFYGISTKFDKQ 182
E F ++++ W+ S + G + GKF T G FYNDAE DKGIQTSQDA+FY S KFD
Sbjct: 25 ENFDNENWEDTWIYSKHPGKEFGKFVLTPGTFYNDAEADKGIQTSQDARFYAASRKFD-G 83
Query: 183 FSNEGKKLVIQFQVKHEQTIDCGGGYVNI 269
FSNE K LV+QF VKHEQ IDCGGGYV +
Sbjct: 84 FSNEDKPLVVQFSVKHEQNIDCGGGYVKL 112
>sp|Q06814|CRTC_SCHMA Calreticulin precursor (SM4 protein) Length = 393 Score = 341 bits (875), Expect = 2e-93 Identities = 158/256 (61%), Positives = 191/256 (74%), Gaps = 5/256 (1%) Frame = +1 Query: 268 LMSSDIDQTKFHGETPYHIMFGPDICGLSTKKVHVIFNYKGQNHLIKKEIRCKDDEFSHV 447 L+ SDID KFHGE+PY IMFGPDICG++TKKVHVIFNYKG+NHLIKKEI CKDD +H+ Sbjct: 110 LLGSDIDPKKFHGESPYKIMFGPDICGMATKKVHVIFNYKGKNHLIKKEIPCKDDLKTHL 169 Query: 448 YTLIVSPDNTYEVLIDNEKVESGNLEDDWDFLAPKKIKDPNAKKPEDWEDNEKIDDSTDK 627 YTLIV+P+N YEVL+DN KVE G+LEDDWD L PKKI DPN KKP+DW D + IDD DK Sbjct: 170 YTLIVNPNNKYEVLVDNAKVEEGSLEDDWDMLPPKKIDDPNDKKPDDWVDEQFIDDPDDK 229 Query: 628 KPEDWDKPKTIADPKAKKPXXXXXXXXXXXXAPQIDNPEYKGEWKPKQIDNPKYKGPWIH 807 KP++WD+PKTI D AKKP PQ DNPEYKGEW P++IDNPKYKG W Sbjct: 230 KPDNWDQPKTIPDMDAKKPDDWDDAMDGEWERPQKDNPEYKGEWTPRRIDNPKYKGEWKP 289 Query: 808 PEIDNPDYKPESNLYLYKNIGSIGFDLWQVKSGTIFDNILLTDDPAYAKKYVEKTWK--- 978 +IDNP+YK + LY+ +IG +GFDLWQV SG+IFDNIL+TD P +AK+ E+ W+ Sbjct: 290 VQIDNPEYKHDPELYVLNDIGYVGFDLWQVDSGSIFDNILITDSPDFAKEEGERLWRKRY 349 Query: 979 --ETAAGEKKMKDKQD 1020 E A + KD ++ Sbjct: 350 DAEVAKEQSSAKDDKE 365
Score = 87.4 bits (215), Expect = 8e-17
Identities = 41/89 (46%), Positives = 59/89 (66%)
Frame = +3
Query: 3 EEFLTDSYKTQWVESTYKGADQGKFTWTAGKFYNDAEKDKGIQTSQDAKFYGISTKFDKQ 182
E F +S + WV+STY QG+F AGK D +D G++T+QDA+FYGI+ K +
Sbjct: 23 ETFPNESIEN-WVQSTYNAEKQGEFKVEAGKSPVDPIEDLGLKTTQDARFYGIARKISEP 81
Query: 183 FSNEGKKLVIQFQVKHEQTIDCGGGYVNI 269
FSN GK +V+QF VK ++T+ CGG Y+ +
Sbjct: 82 FSNRGKTMVLQFTVKFDKTVSCGGAYIKL 110
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 128,573,713
Number of Sequences: 369166
Number of extensions: 2574593
Number of successful extensions: 8654
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 7576
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 8151
length of database: 68,354,980
effective HSP length: 114
effective length of database: 47,295,190
effective search space used: 15087165610
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail