DrC_00581
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00581 (1093 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P25235|RIB2_RAT Dolichyl-diphosphooligosaccharide--prote... 183 7e-46 sp|Q9DBG6|RPN2_MOUSE Dolichyl-diphosphooligosaccharide--pro... 182 1e-45 sp|P04844|RIB2_HUMAN Dolichyl-diphosphooligosaccharide--pro... 182 1e-45 sp|Q6BS08|ATG1_DEBHA Serine/threonine-protein kinase ATG1 (... 41 0.005 sp|Q02795|OSTD_YEAST Dolichyl-diphosphooligosaccharide--pro... 35 0.38 sp|O68433|LEP4_LEGPN Type 4 prepilin-like proteins leader p... 35 0.38 sp|P50976|PTLCB_STRMU PTS system lactose-specific EIICB com... 32 4.2 sp|Q19537|RFA1_CAEEL Probable replication factor A 73 kDa s... 32 4.2 sp|P41003|SMC2_SCHPO Structural maintenance of chromosome 2... 31 7.1 sp|O67151|GCSH1_AQUAE Glycine cleavage system H protein 1 30 9.3
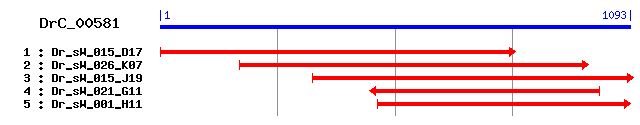
>sp|P25235|RIB2_RAT Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 63 kDa subunit precursor (Ribophorin II) (RPN-II) Length = 631 Score = 183 bits (465), Expect = 7e-46 Identities = 121/351 (34%), Positives = 186/351 (52%), Gaps = 2/351 (0%) Frame = +2 Query: 2 AALVAETVKNVQSATTVLSRKSLVFDRTTGIYELDLTSVQEQLAKGFYSIKLDMKTNETR 181 AA+ E K+V S TVL + + F ++EL+ +V+ L G+Y + ++ + R Sbjct: 303 AAVKLEHAKSVASRATVLQK--MPFSLVGDVFELNFKNVK--LPSGYYDFSVRVE-GDNR 357 Query: 182 YNEAVGVALVVKVICQVSIENVELSIKERDQITGILNYKLTMGSLLSDKLSMNWHHRLDM 361 Y A V L VK+ +V I NV+LS ++DQ ++T + + H + Sbjct: 358 YI-ANTVELRVKISTEVGITNVDLSTVDKDQSIAPKTTRVTYPAKAKGTFIADSHQNFAL 416 Query: 362 KFVIRDHSQSVK--PHQVFVKLTHFNTKQEIIFVVEADRTDVYKFNLDLDKSNKDFDNLS 535 F + D + + PHQ FV+L + T QE++FV E D +VYKF LD + +FD+ S Sbjct: 417 FFQLVDVNTGAELTPHQTFVRLHNQKTGQEVVFVAEPDNKNVYKFELDTSERKIEFDSAS 476 Query: 536 GDYKMELVVGDSMIETPGNWIFANIKLDMPSSPSTENTGESRRTVAGSKRSVVNPRIGSG 715 G Y + L++GD+ ++ P W A++ + P E+ TV +++ P Sbjct: 477 GTYTLYLIIGDATLKNPILWNVADVVIKFPEE-------EAPSTVLS--QNLFTP----- 522 Query: 716 PSLVKPQIFHMFKPAEKRGGQLISTVFTILCIVPPFILMVALWLMVGVNVSNFSVGLSPI 895 K +I H+F+ EKR ++S FT L I+ P +L+ ALW+ +G NVSNF+ S + Sbjct: 523 ----KQEIQHLFREPEKRPPTVVSNTFTAL-ILSPLLLLFALWIRIGANVSNFTFAPSTV 577 Query: 896 MFHVGIIAIFGLYYCYWGYLNMFTTLQCLLPLGMFTAFFGNRVLRQIASKR 1048 +FH+G A+ GL Y YW LNMF TL+ L LG T GNR+L Q A KR Sbjct: 578 IFHLGHAAMLGLMYVYWTQLNMFQTLKYLAVLGTVTFLAGNRMLAQQAVKR 628
>sp|Q9DBG6|RPN2_MOUSE Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 63 kDa subunit precursor (Ribophorin II) (RPN-II) Length = 631 Score = 182 bits (463), Expect = 1e-45 Identities = 120/351 (34%), Positives = 186/351 (52%), Gaps = 2/351 (0%) Frame = +2 Query: 2 AALVAETVKNVQSATTVLSRKSLVFDRTTGIYELDLTSVQEQLAKGFYSIKLDMKTNETR 181 AA+ E K+ + TVL + F ++EL+ +V+ L+ G+Y + ++ ++R Sbjct: 303 AAVKLEHAKSAATRATVLQKTP--FSLVGNVFELNFKNVK--LSSGYYDFSVRVE-GDSR 357 Query: 182 YNEAVGVALVVKVICQVSIENVELSIKERDQITGILNYKLTMGSLLSDKLSMNWHHRLDM 361 Y A V L VK+ +V I NV+LS ++DQ ++T + + H + Sbjct: 358 YI-ANTVELRVKISTEVGITNVDLSTVDKDQSIAPKTTRVTYPAKAKGTFIADSHQNFAL 416 Query: 362 KFVIRDHSQSVK--PHQVFVKLTHFNTKQEIIFVVEADRTDVYKFNLDLDKSNKDFDNLS 535 F + D + + PHQ FV+L + T QE++FV E D +VYKF LD + +FD+ S Sbjct: 417 FFQLVDVNTGAELTPHQTFVRLHNQKTGQEVVFVAEPDNKNVYKFELDTSERKIEFDSAS 476 Query: 536 GDYKMELVVGDSMIETPGNWIFANIKLDMPSSPSTENTGESRRTVAGSKRSVVNPRIGSG 715 G Y + L++GD+ ++ P W A++ + P E+ TV +S+ P Sbjct: 477 GTYTLYLIIGDATLKNPILWNVADVVIKFPEE-------EAPSTVLS--QSLFTP----- 522 Query: 716 PSLVKPQIFHMFKPAEKRGGQLISTVFTILCIVPPFILMVALWLMVGVNVSNFSVGLSPI 895 K +I H+F+ EKR ++S FT L I+ P +L+ ALW+ +G NVSNF+ S + Sbjct: 523 ----KQEIQHLFREPEKRPPTVVSNTFTAL-ILSPLLLLFALWIRIGANVSNFTFAPSTV 577 Query: 896 MFHVGIIAIFGLYYCYWGYLNMFTTLQCLLPLGMFTAFFGNRVLRQIASKR 1048 +FH+G A+ GL Y YW LNMF TL+ L LG T GNR+L Q A KR Sbjct: 578 IFHLGHAAMLGLMYIYWTQLNMFQTLKYLAVLGTVTFLAGNRMLAQHAVKR 628
>sp|P04844|RIB2_HUMAN Dolichyl-diphosphooligosaccharide--protein glycosyltransferase 63 kDa subunit precursor (Ribophorin II) (RPN-II) (RIBIIR) Length = 631 Score = 182 bits (463), Expect = 1e-45 Identities = 121/351 (34%), Positives = 186/351 (52%), Gaps = 2/351 (0%) Frame = +2 Query: 2 AALVAETVKNVQSATTVLSRKSLVFDRTTGIYELDLTSVQEQLAKGFYSIKLDMKTNETR 181 A + E K+V S TVL + S F ++EL+ +V+ + G+Y ++++ + R Sbjct: 303 ATVKLEHAKSVASRATVLQKTS--FTPVGDVFELNFMNVK--FSSGYYDFLVEVE-GDNR 357 Query: 182 YNEAVGVALVVKVICQVSIENVELSIKERDQITGILNYKLTMGSLLSDKLSMNWHHRLDM 361 Y A V L VK+ +V I NV+LS ++DQ ++T + + H + Sbjct: 358 YI-ANTVELRVKISTEVGITNVDLSTVDKDQSIAPKTTRVTYPAKAKGTFIADSHQNFAL 416 Query: 362 KFVIRDHSQSVK--PHQVFVKLTHFNTKQEIIFVVEADRTDVYKFNLDLDKSNKDFDNLS 535 F + D + + PHQ FV+L + T QE++FV E D +VYKF LD + +FD+ S Sbjct: 417 FFQLVDVNTGAELTPHQTFVRLHNQKTGQEVVFVAEPDNKNVYKFELDTSERKIEFDSAS 476 Query: 536 GDYKMELVVGDSMIETPGNWIFANIKLDMPSSPSTENTGESRRTVAGSKRSVVNPRIGSG 715 G Y + L++GD+ ++ P W A++ + P E+ TV +++ P Sbjct: 477 GTYTLYLIIGDATLKNPILWNVADVVIKFPEE-------EAPSTVLS--QNLFTP----- 522 Query: 716 PSLVKPQIFHMFKPAEKRGGQLISTVFTILCIVPPFILMVALWLMVGVNVSNFSVGLSPI 895 K +I H+F+ EKR ++S FT L I+ P +L+ ALW+ +G NVSNF+ S I Sbjct: 523 ----KQEIQHLFREPEKRPPTVVSNTFTAL-ILSPLLLLFALWIRIGANVSNFTFAPSTI 577 Query: 896 MFHVGIIAIFGLYYCYWGYLNMFTTLQCLLPLGMFTAFFGNRVLRQIASKR 1048 +FH+G A+ GL Y YW LNMF TL+ L LG T GNR+L Q A KR Sbjct: 578 IFHLGHAAMLGLMYVYWTQLNMFQTLKYLAILGSVTFLAGNRMLAQQAVKR 628
>sp|Q6BS08|ATG1_DEBHA Serine/threonine-protein kinase ATG1 (Autophagy-related protein 1) Length = 875 Score = 41.2 bits (95), Expect = 0.005 Identities = 36/123 (29%), Positives = 59/123 (47%), Gaps = 9/123 (7%) Frame = +2 Query: 80 RTTGIYELDLTSVQEQLAKGFYSIKLDMKTNETRYNEAVGVALVV--KVICQVSIENVEL 253 +T G+Y S+ ++ KG ++ K T+ NE+V + VV K+ + +EN+E+ Sbjct: 17 KTIGVY-----SIGPEIGKGSFATVY--KCFNTKTNESVAIKSVVRSKLKSKKLVENLEI 69 Query: 254 SIK-----ERDQITGILNYKLTMG--SLLSDKLSMNWHHRLDMKFVIRDHSQSVKPHQVF 412 I + I G+L+YK T L+ D SM D+ + IR +Q +K H V Sbjct: 70 EISILKTMKHPHIVGLLDYKQTTSHFHLVMDYCSMG-----DLSYFIRKRNQLIKTHPVI 124 Query: 413 VKL 421 L Sbjct: 125 SSL 127
>sp|Q02795|OSTD_YEAST Dolichyl-diphosphooligosaccharide--protein glycosyltransferase delta subunit precursor (Oligosaccharyl transferase delta subunit) Length = 286 Score = 35.0 bits (79), Expect = 0.38 Identities = 24/93 (25%), Positives = 43/93 (46%), Gaps = 3/93 (3%) Frame = +2 Query: 725 VKPQIFHMFKPAEKRGGQLISTVFTILCIVPPFILMVALWLMVGVNVSNFSVGLSPIMFH 904 +KP+I H+F KR + I+ +F ++ + L+V +N G++ + F Sbjct: 176 IKPEIHHIFHAEPKRVAKPIAVIFVLIIFITILSLIVTWLNSCAAAFNNIPTGVTAVYFL 235 Query: 905 VGIIAIFG---LYYCYWGYLNMFTTLQCLLPLG 994 I I G ++ Y+ ++F TL L LG Sbjct: 236 GFIATIVGFEVIFARYYLGTSIFETLFSSLYLG 268
>sp|O68433|LEP4_LEGPN Type 4 prepilin-like proteins leader peptide processing enzyme [Includes: Leader peptidase (Prepilin peptidase); N-methyltransferase ] Length = 287 Score = 35.0 bits (79), Expect = 0.38 Identities = 24/84 (28%), Positives = 40/84 (47%), Gaps = 7/84 (8%) Frame = +2 Query: 779 LISTVFTILC--IVPPFILMVALWLMVGVNVSNFSVGLSPIMFHVGIIAIFGLYYCYWGY 952 LIS VF L ++P + + LW+ + N N V L V +++ G Y W + Sbjct: 141 LISLVFIDLDHQLLPDSLTLGLLWIGLIANTQNVFVSLD-----VAVLSCAGAYLALWLF 195 Query: 953 LNMFTTLQCLLPLG-----MFTAF 1009 +N+F + C + +G +F AF Sbjct: 196 INLFYLMTCKVCMGHGDFKLFAAF 219
>sp|P50976|PTLCB_STRMU PTS system lactose-specific EIICB component (EIICB-Lac) (EII-Lac) [Includes: Lactose permease IIC component (PTS system lactose-specific EIIC component); Lactose-specific phosphotransferase enzyme IIB component (PTS system lactose-specific EIIB component)] Length = 568 Score = 31.6 bits (70), Expect = 4.2 Identities = 24/83 (28%), Positives = 42/83 (50%), Gaps = 6/83 (7%) Frame = +2 Query: 716 PSLVKPQIFHMFKPAEKRGGQLISTVFTILCIVPPFILMVALWLMVGVNVSNFSVG--LS 889 P V P I +FK + F+I+ + + + +A+ ++GVNV+ S+G L+ Sbjct: 166 PEEVPPNISQVFK-------DIFPFAFSIIIL---YAIQLAIKAVIGVNVAQ-SIGTLLA 214 Query: 890 PIMF----HVGIIAIFGLYYCYW 946 P+ ++GI IFG Y +W Sbjct: 215 PLFSAADGYLGITIIFGAYALFW 237
>sp|Q19537|RFA1_CAEEL Probable replication factor A 73 kDa subunit (RP-A) (RF-A) (Replication factor-A protein 1) Length = 655 Score = 31.6 bits (70), Expect = 4.2 Identities = 18/64 (28%), Positives = 31/64 (48%) Frame = +2 Query: 413 VKLTHFNTKQEIIFVVEADRTDVYKFNLDLDKSNKDFDNLSGDYKMELVVGDSMIETPGN 592 ++ T FN E ++ + Y + ++NK F+N DY++ L DS+IE G Sbjct: 271 IRCTAFNEVAESLYTTITENLSYYLSGGSVKQANKKFNNTGHDYEITL-RSDSIIEAGGE 329 Query: 593 WIFA 604 + A Sbjct: 330 LLAA 333
>sp|P41003|SMC2_SCHPO Structural maintenance of chromosome 2 (Chromosome segregation protein cut14) (Cell untimely torn protein 14) Length = 1172 Score = 30.8 bits (68), Expect = 7.1 Identities = 55/277 (19%), Positives = 109/277 (39%), Gaps = 15/277 (5%) Frame = +2 Query: 17 ETVKNVQSATTVLSRKSLVFDRTTGIYELDLT-------SVQEQLAKGF--YSIKLDMKT 169 + VK+++ L R L D+ E D+ S +L K F Y KLD T Sbjct: 751 DDVKDLKQRLPELDRLILQSDQAIKKIERDMQEWKHNKGSKMAELEKEFNQYKHKLDEFT 810 Query: 170 N--ETRYNEAVGVALVVKVICQVSIENVELSIKERDQITGILNYKLTMGSLLSDKLSMNW 343 E N+ GV L + + + ++N + S+ + + T ++ ++ Sbjct: 811 PILEKSENDYNGVKLECEQL-EGELQNHQQSLVQGESTTSLIKTEIA------------- 856 Query: 344 HHRLDMKFVIRDHSQSVKPHQVFVKLTHFN-TKQEIIFVVEADRTDVYKFN---LDLDKS 511 L++ V +H++ + ++ F+ +EI + + +T + N L + K Sbjct: 857 --ELELSLVNEEHNRKKLTELIEIESAKFSGLNKEIDSLSTSMKTFESEINNGELTIQKL 914 Query: 512 NKDFDNLSGDYKMELVVGDSMIETPGNWIFANIKLDMPSSPSTENTGESRRTVAGSKRSV 691 N +FD L + K + + +E +WI + + ++ R ++ Sbjct: 915 NHEFDRLERE-KSVAITAINHLEKENDWIDGQKQHFGKQGTIFDFHSQNMRQCREQLHNL 973 Query: 692 VNPRIGSGPSLVKPQIFHMFKPAEKRGGQLISTVFTI 802 PR S + P++ M EK+ +L S + TI Sbjct: 974 -KPRFASMRKAINPKVMDMIDGVEKKEAKLRSMIKTI 1009
>sp|O67151|GCSH1_AQUAE Glycine cleavage system H protein 1 Length = 143 Score = 30.4 bits (67), Expect = 9.3 Identities = 15/44 (34%), Positives = 23/44 (52%) Frame = +2 Query: 71 VFDRTTGIYELDLTSVQEQLAKGFYSIKLDMKTNETRYNEAVGV 202 V D GIY + + S+ LA YSIK+ + Y+EA+ + Sbjct: 19 VKDEGNGIYSVGMASILAALAYPLYSIKIKPVGTKLEYDEALAI 62
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 135,174,053
Number of Sequences: 369166
Number of extensions: 2966107
Number of successful extensions: 8264
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 7787
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 8236
length of database: 68,354,980
effective HSP length: 112
effective length of database: 47,664,660
effective search space used: 11963829660
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail