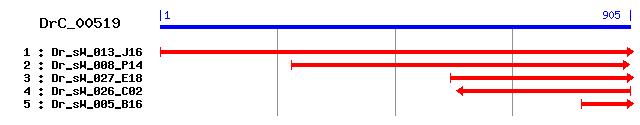
DrC_00519
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00519 (905 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q37376|NU2M_ACACA NADH-ubiquinone oxidoreductase chain 2... 50 9e-06 sp|O99817|NU2M_RHISA NADH-ubiquinone oxidoreductase chain 2... 48 4e-05 sp|P24499|ATP6_TRYBB ATP synthase a chain (ATPase protein 6) 34 0.64 sp|P48902|NU2M_ALBCO NADH-ubiquinone oxidoreductase chain 2... 33 0.84 sp|P15577|NU2M_PARTE NADH-ubiquinone oxidoreductase chain 2... 32 1.9 sp|P18931|NU4M_DROME NADH-ubiquinone oxidoreductase chain 4... 32 1.9 sp|P56911|NUON2_RHIME NADH-quinone oxidoreductase chain N 2... 31 5.4 sp|O47478|NU6M_LOLBL NADH-ubiquinone oxidoreductase chain 6... 31 5.4 sp|O24982|LNT_HELPY Apolipoprotein N-acyltransferase (ALP N... 31 5.4 sp|P26361|CFTR_MOUSE Cystic fibrosis transmembrane conducta... 30 7.1
>sp|Q37376|NU2M_ACACA NADH-ubiquinone oxidoreductase chain 2 (NADH dehydrogenase subunit 2) Length = 527 Score = 50.1 bits (118), Expect = 9e-06 Identities = 68/285 (23%), Positives = 109/285 (38%), Gaps = 28/285 (9%) Frame = +2 Query: 107 IIHYFGGLSYNINSLF----YILAVLL-SDILVMSGFIVVSFDYLFFGFMLKLSFFPFLW 271 I + G LSY +LF Y+ +L S SGF++ F L GF+ KL PF Sbjct: 212 IYGFTGLLSYTDLTLFLSEVYVTNFILDSSFFSFSGFLI-GFLLLTVGFLFKLGSAPFHM 270 Query: 272 LVPNLFLGMSYWLIGFLVFFHKAVIVXXXXXXXXXXXXXHVFLILLSIF-VSRLYLILNK 448 +P+++ G + +L K ++ VFL +F ++ L+ IL Sbjct: 271 WMPDVYEGSPLLITAYLSTLPKISLIFVIFKLYYYVFF-EVFLFSQGLFTLTALFSILLG 329 Query: 449 DC-------LKSLIIWSSNIHCGXXXXXXXXXXXXVFGYMLIYLIFGIAFLIYLCYSF-- 601 LK ++ +S + G + YLI I +I L + F Sbjct: 330 SIAAIYQVKLKRMLTYSMITNTGYLLLGLSFGDISGIYITIFYLISYIFIMIGLFFCFLS 389 Query: 602 -----------EYNYFSDVVGLDNGYGXXXXXXXXXXCRFPPMLG*VLKFTVYSTILKYT 748 N FS+++ ++ PP+LG KF ++ LKY Sbjct: 390 LRDRSSGLLVKRINLFSNLLEVNPSLSFSIFILLFSIAGIPPLLGFYSKFFLFLFSLKYK 449 Query: 749 ILFFLILVNLFSTFGYLVVLRSFFVLLPY*NHFK--IYFYSLPFI 877 + + IL +FS +R V L Y N I+ Y +PF+ Sbjct: 450 MYWMTILFVIFSVVSVFYYIR--LVKLMYFNRTTGWIFLYDIPFL 492
>sp|O99817|NU2M_RHISA NADH-ubiquinone oxidoreductase chain 2 (NADH dehydrogenase subunit 2) Length = 313 Score = 47.8 bits (112), Expect = 4e-05 Identities = 50/256 (19%), Positives = 97/256 (37%), Gaps = 11/256 (4%) Frame = +2 Query: 92 ISLFAIIHYFGGLSYNINSLFYILAVLLSDILVMSGFIVVSFDYLFFGFMLKLSFFPFLW 271 +S I++ F +YN ++I+ S + F + SF Y F L S Sbjct: 31 MSFIPIMNDFKMKNYNSMITYFIIQSFSSSL-----FFISSFQYSLFNMELFFSLINISV 85 Query: 272 LVPNLFLGMSYWLIGF---LVFFHKAVIVXXXXXXXXXXXXXHVFLILLSIFVSRLYL-- 436 L+ + +WLI L F+ +++ + +++ +F+ L Sbjct: 86 LIKLGMIPFHFWLIMISESLTFYSLLILLTIQKIIPLLIIEKFMNSLMIPLFIMSSLLAS 145 Query: 437 --ILNKDCLKSLIIWSSNIHCGXXXXXXXXXXXXVFGYMLIYLIFGIAFLIYLCYSFEYN 610 + +K ++I+SS H G Y+++Y I I +I C +N Sbjct: 146 IMAMKYKLIKQILIFSSISHQGWILCLIAKKMNFWVSYLMLYSII-IYSIIINCKEMNFN 204 Query: 611 YFSDVV--GLDNGYGXXXXXXXXXXCRFPPMLG*VLKFTVYSTILKYTILFFLILV--NL 778 D+ + + PP +G +K ++K ++F +IL+ +L Sbjct: 205 KLCDLTKKKISSKMKNTMIMSMMSLAGMPPFIGFFMKIMAIFFLMKMNLIFMMILIISSL 264 Query: 779 FSTFGYLVVLRSFFVL 826 + F YL +L F + Sbjct: 265 INLFFYLRILTPLFFI 280
>sp|P24499|ATP6_TRYBB ATP synthase a chain (ATPase protein 6) Length = 229 Score = 33.9 bits (76), Expect = 0.64 Identities = 29/116 (25%), Positives = 50/116 (43%), Gaps = 1/116 (0%) Frame = +2 Query: 542 YMLIYLIFGIAFLIYLCYSFEYNYFSDVVGLDNGYGXXXXXXXXXXCRFPPMLG*VLKF- 718 Y+ +YL+F I FL+Y+ + F + + D L N RF +L F Sbjct: 78 YLNLYLLFCIVFLLYIAFLFLFCFLCDFF-LFNNLLVGDSFMDVFFIRF------LLCFL 130 Query: 719 TVYSTILKYTILFFLILVNLFSTFGYLVVLRSFFVLLPY*NHFKIYFYSLPFIIVF 886 +S + + F + NL S+ L++ FF + + ++ Y + FI VF Sbjct: 131 ECFSLLCRCLSTFLRLFCNLLSSHFLLLMFFDFFYFIFVFFFYGVFCYFILFIFVF 186
>sp|P48902|NU2M_ALBCO NADH-ubiquinone oxidoreductase chain 2 (NADH dehydrogenase subunit 2) Length = 307 Score = 33.5 bits (75), Expect = 0.84 Identities = 54/286 (18%), Positives = 110/286 (38%), Gaps = 19/286 (6%) Frame = +2 Query: 92 ISLFAIIHYFGGLSYNINS----LFYILAVLLSDILVMSGFIVV-----SFDYLFFGF-- 238 ISL + Y+ + ++ +++++ + S +++++G + S+ YLF Sbjct: 34 ISLLGFVSYYMLMKKIMSGEGIMMYFLIQSVSSTVMLLNGLYIFVNHASSYIYLFIFITM 93 Query: 239 -MLKLSFFPF-LWLVPNLFLGMSYWLIGFLVFFHKAVIVXXXXXXXXXXXXXHVFLILLS 412 MLK+ FP W++P ++ +SY IG + K V + + +LS Sbjct: 94 LMLKIGMFPLHFWIIP-VYSKLSYLNIGIVGLLLKIVPMWILMHMGCITSEMLNLITMLS 152 Query: 413 IFVSRLYLILNKDCLKSLIIWSSNIHCGXXXXXXXXXXXXVFGYMLIYLIFGIAFLIYLC 592 + ++ + K ++ ++ +F Y + Y + L++L Sbjct: 153 VTSMLFGALIGMNLSKMRMVLGASTITHNGWLGMSCISGSLFKYFITYGFSLVILLVFLY 212 Query: 593 YSFEYNYFSDVVGLDNGYGXXXXXXXXXXCRFPPMLG*VLKFTV----YSTILKYTILFF 760 + + ++ L PP + + K V T L + +L F Sbjct: 213 LGDKMSISLSLLSLSG---------------LPPFMLFIGKINVLLMMMETNLWFIVLVF 257 Query: 761 LILVNLFSTFGYLVVLRSFFVLLP--Y*NHFKIYFYSLPFIIVFFG 892 IL + S YL FF+ + Y H+K+ + L + V FG Sbjct: 258 AILSAVISLVYYLKFSVMFFMNMKNNYLKHYKMAMFLL--VNVTFG 301
>sp|P15577|NU2M_PARTE NADH-ubiquinone oxidoreductase chain 2 (NADH dehydrogenase subunit 2) Length = 193 Score = 32.3 bits (72), Expect = 1.9 Identities = 25/86 (29%), Positives = 42/86 (48%), Gaps = 14/86 (16%) Frame = +2 Query: 689 PPMLG*VLKFTVYSTILKYTILFFLIL---VNLFSTFGYLVVLRSFF-----VLLPY*NH 844 PP+LG LKF ++ + T L F+++ N+ + F YL ++SF +L N Sbjct: 96 PPLLGFFLKFLIFFFLFFKTNLAFILIFLGFNMATLFFYLSTVKSFVNRKQASVLNSFNF 155 Query: 845 F------KIYFYSLPFIIVFFG*FLL 904 F +YF++ + +FF F L Sbjct: 156 FIRAELSFLYFFNFFYFFLFFAFFFL 181
>sp|P18931|NU4M_DROME NADH-ubiquinone oxidoreductase chain 4 (NADH dehydrogenase subunit 4) Length = 446 Score = 32.3 bits (72), Expect = 1.9 Identities = 20/78 (25%), Positives = 37/78 (47%), Gaps = 8/78 (10%) Frame = +2 Query: 239 MLKLSFFPFLWLVPNLFLGMSYWLIGFLVFFHKAVIVXXXXXXXXXXXXXHVF------- 397 MLK+ FF L+L+P F+ YW++ ++FF + + + Sbjct: 1 MLKIIFF-LLFLIPFCFINNMYWMVQIMMFFISFIFLLMNNFMNYWSEISYFLGCDMLSY 59 Query: 398 -LILLSIFVSRLYLILNK 448 LILLS+++ L L+ ++ Sbjct: 60 GLILLSLWICSLMLLASE 77
>sp|P56911|NUON2_RHIME NADH-quinone oxidoreductase chain N 2 (NADH dehydrogenase I, chain N 2) (NDH-1, chain N 2) Length = 479 Score = 30.8 bits (68), Expect = 5.4 Identities = 45/231 (19%), Positives = 87/231 (37%), Gaps = 21/231 (9%) Frame = +2 Query: 203 IVVSFDYLFFGFMLKLSFFPFLWLVPNLFLGMSYWLIGFLVFFHKAV--IVXXXXXXXXX 376 ++++ ++ G + K+ PF VP+++ G + F+ KA V Sbjct: 206 LLLAVVFIVAGLVFKIGAVPFHMWVPDVYEGAPTTITAFMSVAPKAAGFAVILRVFLNPL 265 Query: 377 XXXXHVFLILLSIFVSRL----YLILNKDCLKSLIIWSSNIHCG----XXXXXXXXXXXX 532 + ++I+ +I V+ + ++ L +D K L+ +SS H G Sbjct: 266 VEASNAWIIVAAIAVATMALGSFVALVQDNFKRLLAYSSIAHAGFAIFGVVAGGQDGIAS 325 Query: 533 VFGYMLIYL-----IFGIAFLIYLCYSFEYNYFSDVVGLDN---GYGXXXXXXXXXXCRF 688 V Y+LIY IFG A ++ F D G G Sbjct: 326 VMLYLLIYTFMNLGIFG-AVIMMRNGDFSGEVIEDYAGFAKFHPGLALLMLLYLFSLAGI 384 Query: 689 PPMLG*VLKFTVYSTILK---YTILFFLILVNLFSTFGYLVVLRSFFVLLP 832 PP G KF V +++ + +L+++ S + Y+ ++ ++ P Sbjct: 385 PPTAGFFAKFYVLVALVERGFVALAVIAVLLSVVSAYFYIRIVMVIYMREP 435
>sp|O47478|NU6M_LOLBL NADH-ubiquinone oxidoreductase chain 6 (NADH dehydrogenase subunit 6) Length = 168 Score = 30.8 bits (68), Expect = 5.4 Identities = 22/100 (22%), Positives = 45/100 (45%) Frame = +2 Query: 116 YFGGLSYNINSLFYILAVLLSDILVMSGFIVVSFDYLFFGFMLKLSFFPFLWLVPNLFLG 295 Y GG+ +F + L+ +++ +S + F ++FFGFM+ ++FF LV Sbjct: 57 YVGGMLV----MFMYVISLIPNLIFLSNKVFAYFFFIFFGFMM-MNFFVMKELVSVEVKS 111 Query: 296 MSYWLIGFLVFFHKAVIVXXXXXXXXXXXXXHVFLILLSI 415 MS + G++ +I+ + +L+S+ Sbjct: 112 MSLFDYGYMSMGGSGIIMLYDNFFCYVLLAVILLFVLISV 151
>sp|O24982|LNT_HELPY Apolipoprotein N-acyltransferase (ALP N-acyltransferase) Length = 425 Score = 30.8 bits (68), Expect = 5.4 Identities = 20/78 (25%), Positives = 36/78 (46%), Gaps = 8/78 (10%) Frame = +2 Query: 104 AIIHYFGGLSYNINSLFYILAVLLSDILVMSGFIVVSFDYLFFGFMLKLSF--------F 259 A++ Y+ LS+ + Y+L +++ I ++ G + Y + LSF F Sbjct: 70 ALLFYWCALSFRYSDFTYLLPLIIVLIALVYGVLFYLLLYFENPYFRLLSFLGSSFIHPF 129 Query: 260 PFLWLVPNLFLGMSYWLI 313 F WLVP+ F S + + Sbjct: 130 GFDWLVPDSFFSYSVFRV 147
>sp|P26361|CFTR_MOUSE Cystic fibrosis transmembrane conductance regulator (CFTR) (cAMP-dependent chloride channel) Length = 1476 Score = 30.4 bits (67), Expect = 7.1 Identities = 11/32 (34%), Positives = 22/32 (68%) Frame = +2 Query: 716 FTVYSTILKYTILFFLILVNLFSTFGYLVVLR 811 F V+ ++L YT++ ++L +F+T + +VLR Sbjct: 316 FVVFLSVLPYTVINGIVLRKIFTTISFCIVLR 347
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 67,411,572
Number of Sequences: 369166
Number of extensions: 1061026
Number of successful extensions: 3346
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 3146
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 3334
length of database: 68,354,980
effective HSP length: 110
effective length of database: 48,034,130
effective search space used: 9174518830
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail