DrC_00464
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
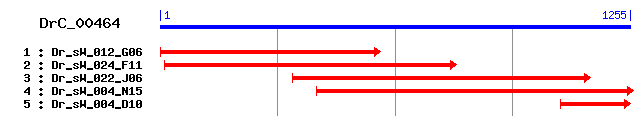
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00464 (1255 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q00341|VIGLN_HUMAN Vigilin (High density lipoprotein-bin... 204 6e-52 sp|P81021|VIGLN_CHICK Vigilin 203 7e-52 sp|Q8VDJ3|VIGLN_MOUSE Vigilin (High density lipoprotein-bin... 202 1e-51 sp|Q9Z1A6|VIGLN_RAT Vigilin (High density lipoprotein-bindi... 200 8e-51 sp|P06105|SC160_YEAST Protein SCP160 (Protein HX) 79 2e-14 sp|Q80VL1|TDRKH_MOUSE Tudor and KH domain-containing protein 43 0.002 sp|Q9Y2W6|TDRKH_HUMAN Tudor and KH domain-containing protein 42 0.005 sp|P57722|PCBP3_MOUSE Poly(rC)-binding protein 3 (Alpha-CP3) 41 0.008 sp|P57721|PCBP3_HUMAN Poly(rC)-binding protein 3 (Alpha-CP3) 41 0.008 sp|Q01570|SECA_PAVLU Preprotein translocase secA subunit 39 0.031
>sp|Q00341|VIGLN_HUMAN Vigilin (High density lipoprotein-binding protein) (HDL-binding protein) Length = 1268 Score = 204 bits (518), Expect = 6e-52 Identities = 133/392 (33%), Positives = 209/392 (53%), Gaps = 11/392 (2%) Frame = +1 Query: 4 QKHHRNLITRNMTEIKNIQNSFDVMIKFPNRDISS-KSGENGYDEAHLNGNSDNTFVDTT 180 QK HR+++ + I+ I F V IKFP+R+ ++ S E E NG D Sbjct: 882 QKFHRSVMGPKGSRIQQITRDFSVQIKFPDREENAVHSTEPVVQE---NG-------DEA 931 Query: 181 EEGDSNVEAEQNNP--ANWVALSGRRENCESAKEALLELVPVSEDVPLNNEFHRNFIGQK 354 EG + + +P + + +SGR+E CE+AKEAL LVPV+ +V + + HR IGQK Sbjct: 932 GEGREAKDCDPGSPRRCDIIIISGRKEKCEAAKEALEALVPVTIEVEVPFDLHRYVIGQK 991 Query: 355 GQGINTFSTKYNVVVNIPPANKCLDFISIKGPRANLIDAKAGLDELIVVWENEKQDRIAR 534 G GI ++ V +++P D I+I G ANL AKAGL E + + E++DR R Sbjct: 992 GSGIRKMMDEFEVNIHVPAPELQSDIIAITGLAANLDRAKAGLLERVKELQAEQEDRALR 1051 Query: 535 NFENQVPVPIVFHRKLIGQRGANIEMFRSKFNVSIQIPDQ---NSKSEEITIRGYEEDVK 705 +F+ V V +H K+IG++GA I R + +V+IQ PD+ N ++ITI GYE++ + Sbjct: 1052 SFKLSVTVDPKYHPKIIGRKGAVITQIRLEHDVNIQFPDKDDGNQPQDQITITGYEKNTE 1111 Query: 706 KAEDELLSMVDRWKAQVIEELHIPRDVHPQLIGSRGSALRKLREQYNVEINFPSKNVDDD 885 A D +L +V + V E++ + VH ++IG+RG A+RK+ +++ V+I FP D Sbjct: 1112 AARDAILRIVGELEQMVSEDVPLDHRVHARIIGARGKAIRKIMDEFKVDIRFPQSGAPDP 1171 Query: 886 NANRVEVIGDKENVDKCCDILVEKANELLYEWEERNQVREVKNSIKDRENVNSNNLKEKG 1065 N V V G ENV++ D ++ E L + + ++ E K Sbjct: 1172 NC--VTVTGLPENVEEAIDHILNLEEEYLADVVDSEALQVYMKPPAHEE------AKAPS 1223 Query: 1066 PGFLLAGAPW-----QRVPDASSNVDFPTIDA 1146 GF++ APW ++ PD SS+ +FP+ A Sbjct: 1224 RGFVVRDAPWTASSSEKAPDMSSSEEFPSFGA 1255
Score = 114 bits (286), Expect = 5e-25
Identities = 84/330 (25%), Positives = 147/330 (44%), Gaps = 8/330 (2%)
Frame = +1
Query: 1 EQKHHRNLITRNMTEIKNIQNSFDVMIKFPNRDISSKSGENGYDEAHLNGNSDNTFVDTT 180
+ K HR+LI ++ I I++ + V ++ P
Sbjct: 443 DHKFHRHLIGKSGANINRIKDQYKVSVRIP------------------------------ 472
Query: 181 EEGDSNVEAEQNNPANWVALSGRRENCESAKEALLELVPVSE-----DVPLNNEFHRNFI 345
+ +N + + G + + AK LLEL E D+ + FHR I
Sbjct: 473 ---------PDSEKSNLIRIEGDPQGVQQAKRELLELASRMENERTKDLIIEQRFHRTII 523
Query: 346 GQKGQGINTFSTKY-NVVVNIPPANKCLDFISIKGPRANLIDAKAGLDELIVVWENEKQD 522
GQKG+ I K+ V++N P + D + ++GP+ + + +++ D
Sbjct: 524 GQKGERIREIRDKFPEVIINFPDPAQKSDIVQLRGPKNEVEKCTKYMQKMVA-------D 576
Query: 523 RIARNFENQVPVPIVFHRKLIGQRGANIEMFRSKFNVSIQIPDQNSKSEEITIRGYEEDV 702
+ ++ VP+ FH+ +IG+ GANI+ R + N I +P +NS SE I I G +
Sbjct: 577 LVENSYSISVPIFKQFHKNIIGKGGANIKKIREESNTKIDLPAENSNSETIIITGKRANC 636
Query: 703 KKAEDELLSMVDRWKAQVIE-ELHIPRDVHPQLIGSRGSALRKLREQY-NVEINFPSKNV 876
+ A +LS + + A + E E+ IP +H LIG++G +R + E+ V I+FP V
Sbjct: 637 EAARSRILS-IQKDLANIAEVEVSIPAKLHNSLIGTKGRLIRSIMEECGGVHIHFP---V 692
Query: 877 DDDNANRVEVIGDKENVDKCCDILVEKANE 966
+ ++ V + G +V+K L+ A E
Sbjct: 693 EGSGSDTVVIRGPSSDVEKAKKQLLHLAEE 722
Score = 103 bits (257), Expect = 1e-21
Identities = 87/308 (28%), Positives = 147/308 (47%), Gaps = 10/308 (3%)
Frame = +1
Query: 37 MTEIKNIQNSFDVMI----KFPNRDISSKSGENGYDEAHLNGNSDNTFVDTTEEGDSNVE 204
+TE+ NSF V + +R I K G+N A + ++ TE G+ +
Sbjct: 354 LTEVYAKANSFTVSSVAAPSWLHRFIIGKKGQN---LAKITQQMPKVHIEFTE-GEDKIT 409
Query: 205 AEQNNPANWVALSGRRENCESAKEALLELVPVSEDVPLNNEFHRNFIGQKGQGINTFSTK 384
E P V ++ +E E + L+ + E + ++++FHR+ IG+ G IN +
Sbjct: 410 LE--GPTEDVNVA--QEQIEGMVKDLINRMDYVE-INIDHKFHRHLIGKSGANINRIKDQ 464
Query: 385 YNVVVNIPPANKCLDFISIKGPRANLIDAKAGLDELIVVWENEKQDRIARNFENQVPVPI 564
Y V V IPP ++ + I I+G + AK L EL ENE+ + +
Sbjct: 465 YKVSVRIPPDSEKSNLIRIEGDPQGVQQAKRELLELASRMENER--------TKDLIIEQ 516
Query: 565 VFHRKLIGQRGANIEMFRSKF-NVSIQIPDQNSKSEEITIRGYEEDVKKAEDELLSMVDR 741
FHR +IGQ+G I R KF V I PD KS+ + +RG + +V+K + MV
Sbjct: 517 RFHRTIIGQKGERIREIRDKFPEVIINFPDPAQKSDIVQLRGPKNEVEKCTKYMQKMV-- 574
Query: 742 WKAQVIEELH-----IPRDVHPQLIGSRGSALRKLREQYNVEINFPSKNVDDDNANRVEV 906
A ++E + I + H +IG G+ ++K+RE+ N +I+ P++N N+ + +
Sbjct: 575 --ADLVENSYSISVPIFKQFHKNIIGKGGANIKKIREESNTKIDLPAEN---SNSETIII 629
Query: 907 IGDKENVD 930
G + N +
Sbjct: 630 TGKRANCE 637
Score = 92.0 bits (227), Expect = 3e-18
Identities = 90/371 (24%), Positives = 165/371 (44%), Gaps = 43/371 (11%)
Frame = +1
Query: 1 EQKHHRNLITRNMTEIKNIQNSF-DVMIKFPN----RDISSKSG---------------- 117
EQ+ HR +I + I+ I++ F +V+I FP+ DI G
Sbjct: 515 EQRFHRTIIGQKGERIREIRDKFPEVIINFPDPAQKSDIVQLRGPKNEVEKCTKYMQKMV 574
Query: 118 ----ENGYD-------EAHLN--GNSDNTFVDTTEEGDSNVEAE-QNNPANWVALSGRRE 255
EN Y + H N G EE ++ ++ +N+ + + ++G+R
Sbjct: 575 ADLVENSYSISVPIFKQFHKNIIGKGGANIKKIREESNTKIDLPAENSNSETIIITGKRA 634
Query: 256 NCESAKEALL----ELVPVSE-DVPLNNEFHRNFIGQKGQGINTFSTKY-NVVVNIPPAN 417
NCE+A+ +L +L ++E +V + + H + IG KG+ I + + V ++ P
Sbjct: 635 NCEAARSRILSIQKDLANIAEVEVSIPAKLHNSLIGTKGRLIRSIMEECGGVHIHFPVEG 694
Query: 418 KCLDFISIKGPRANLIDAKAGLDELIVVWENEKQDRIARNFENQVPVPIVFHRKLIGQRG 597
D + I+GP +++ AK L L EKQ ++F + +H+ LIG+ G
Sbjct: 695 SGSDTVVIRGPSSDVEKAKKQLLHL----AEEKQ---TKSFTVDIRAKPEYHKFLIGKGG 747
Query: 598 ANIEMFRSKFNVSIQIPDQNSKSEE-ITIRGYEEDVKKAEDELLSMVDRWKAQVIEELHI 774
I R + P K ++ ITI G E+ V++A+ EL +++ V + + +
Sbjct: 748 GKIRKVRDSTGARVIFPAAEDKDQDLITIIGKEDAVREAQKELEALIQNLDNVVEDSMLV 807
Query: 775 PRDVHPQLIGSRGSALRKLREQY-NVEINFPSKNVDDDNANRVEVIGDKENVDKCCDILV 951
H + RG LR++ E+Y V ++FP D +V + G K+ C +
Sbjct: 808 DPKHHRHFVIRRGQVLREIAEEYGGVMVSFPRSGTQSD---KVTLKGAKD----CVEAAK 860
Query: 952 EKANELLYEWE 984
++ E++ + E
Sbjct: 861 KRIQEIIEDLE 871
Score = 91.3 bits (225), Expect = 5e-18
Identities = 62/216 (28%), Positives = 113/216 (52%), Gaps = 2/216 (0%)
Frame = +1
Query: 331 HRNFIGQKGQGINTFSTKY-NVVVNIPPANKCLDFISIKGPRANLIDAKAGLDELIVVWE 507
HR IG+KGQ + + + V + D I+++GP ++ A+ ++ ++
Sbjct: 376 HRFIIGKKGQNLAKITQQMPKVHIEFTEGE---DKITLEGPTEDVNVAQEQIEGMV---- 428
Query: 508 NEKQDRIARNFENQVPVPIVFHRKLIGQRGANIEMFRSKFNVSIQIPDQNSKSEEITIRG 687
+D I R ++ + FHR LIG+ GANI + ++ VS++IP + KS I I G
Sbjct: 429 ---KDLINRMDYVEINIDHKFHRHLIGKSGANINRIKDQYKVSVRIPPDSEKSNLIRIEG 485
Query: 688 YEEDVKKAEDELLSMVDRWKAQVIEELHIPRDVHPQLIGSRGSALRKLREQY-NVEINFP 864
+ V++A+ ELL + R + + ++L I + H +IG +G +R++R+++ V INFP
Sbjct: 486 DPQGVQQAKRELLELASRMENERTKDLIIEQRFHRTIIGQKGERIREIRDKFPEVIINFP 545
Query: 865 SKNVDDDNANRVEVIGDKENVDKCCDILVEKANELL 972
D V++ G K V+KC + + +L+
Sbjct: 546 DPAQKSD---IVQLRGPKNEVEKCTKYMQKMVADLV 578
Score = 86.7 bits (213), Expect = 1e-16
Identities = 69/310 (22%), Positives = 143/310 (46%), Gaps = 38/310 (12%)
Frame = +1
Query: 205 AEQNNPANWVALSGRRENCESAKEALLELVP-----VSEDVPLNNEFHRNFIGQKGQGIN 369
A ++ + + + G+ + A++ L L+ V + + ++ + HR+F+ ++GQ +
Sbjct: 765 AAEDKDQDLITIIGKEDAVREAQKELEALIQNLDNVVEDSMLVDPKHHRHFVIRRGQVLR 824
Query: 370 TFSTKYN-VVVNIPPANKCLDFISIKGPRANLIDAKAGLDELIVVWENEKQDRIARNFEN 546
+ +Y V+V+ P + D +++KG + + AK + E+I + +
Sbjct: 825 EIAEEYGGVMVSFPRSGTQSDKVTLKGAKDCVEAAKKRIQEII--------EDLEAQVTL 876
Query: 547 QVPVPIVFHRKLIGQRGANIEMFRSKFNVSIQIPDQNS---------------------- 660
+ +P FHR ++G +G+ I+ F+V I+ PD+
Sbjct: 877 ECAIPQKFHRSVMGPKGSRIQQITRDFSVQIKFPDREENAVHSTEPVVQENGDEAGEGRE 936
Query: 661 ----------KSEEITIRGYEEDVKKAEDELLSMVDRWKAQVIEELHIPRDVHPQLIGSR 810
+ + I I G +E + A++ L ++V V E+ +P D+H +IG +
Sbjct: 937 AKDCDPGSPRRCDIIIISGRKEKCEAAKEALEALV-----PVTIEVEVPFDLHRYVIGQK 991
Query: 811 GSALRKLREQYNVEINFPSKNVDDDNANRVEVIGDKENVDKCCDILVEKANELLYEWEER 990
GS +RK+ +++ V I+ P+ + D + + G N+D+ L+E+ EL E E+R
Sbjct: 992 GSGIRKMMDEFEVNIHVPAPELQSD---IIAITGLAANLDRAKAGLLERVKELQAEQEDR 1048
Query: 991 NQVREVKNSI 1020
+R K S+
Sbjct: 1049 -ALRSFKLSV 1057
Score = 77.0 bits (188), Expect = 1e-13
Identities = 55/228 (24%), Positives = 110/228 (48%), Gaps = 8/228 (3%)
Frame = +1
Query: 211 QNNPANWVALSGRRENCESAKEALLELVPVSE------DVPLNNEFHRNFIGQKGQGINT 372
+ + ++ V + G + E AK+ LL L + D+ E+H+ IG+ G I
Sbjct: 693 EGSGSDTVVIRGPSSDVEKAKKQLLHLAEEKQTKSFTVDIRAKPEYHKFLIGKGGGKIRK 752
Query: 373 FSTKYNVVVNIPPA-NKCLDFISIKGPRANLIDAKAGLDELIVVWENEKQDRIARNFENQ 549
V P A +K D I+I G + +A+ L+ LI +N +D +
Sbjct: 753 VRDSTGARVIFPAAEDKDQDLITIIGKEDAVREAQKELEALIQNLDNVVEDSML------ 806
Query: 550 VPVPIVFHRKLIGQRGANIEMFRSKFN-VSIQIPDQNSKSEEITIRGYEEDVKKAEDELL 726
V HR + +RG + ++ V + P ++S+++T++G ++ V+ A+ +
Sbjct: 807 --VDPKHHRHFVIRRGQVLREIAEEYGGVMVSFPRSGTQSDKVTLKGAKDCVEAAKKRIQ 864
Query: 727 SMVDRWKAQVIEELHIPRDVHPQLIGSRGSALRKLREQYNVEINFPSK 870
+++ +AQV E IP+ H ++G +GS ++++ ++V+I FP +
Sbjct: 865 EIIEDLEAQVTLECAIPQKFHRSVMGPKGSRIQQITRDFSVQIKFPDR 912
Score = 67.4 bits (163), Expect = 8e-11
Identities = 61/292 (20%), Positives = 131/292 (44%), Gaps = 9/292 (3%)
Frame = +1
Query: 214 NNPANWVALSGRRENCESAKEALLELVPVSED------VPLNNEFHRNFIGQKGQGINTF 375
++P+N + ++G +E E A+ +L L+ +D + + FH G + +
Sbjct: 190 DDPSNQIKITGTKEGIEKARHEVL-LISAEQDKRAVERLEVEKAFHPFIAGPYNRLVGEI 248
Query: 376 STKYNVVVNIPPANKCLDFISIKGPRANLIDAKAGLDELIVVWENEKQDRIARNFENQVP 555
+ +NIPP + I G + L A A + + ++E +K+ + V
Sbjct: 249 MQETGTRINIPPPSVNRTEIVFTGEKEQLAQAVARIKK---IYEEKKK----KTTTIAVE 301
Query: 556 VPIVFHRKLIGQRGANIEMFRSKFNVSIQIPDQNSKSEEITIRGYEEDVKKAEDELLSMV 735
V H+ +IG +G +++ + VS++IP +S SE + +RG E + +A E+ +
Sbjct: 302 VKKSQHKYVIGPKGNSLQEILERTGVSVEIPPSDSISETVILRGEPEKLGQALTEVYAKA 361
Query: 736 DRWKAQVIEELHIPRDVHPQLIGSRGSALRKLREQY-NVEINFPSKNVDDDNANRVEVIG 912
+ + + + P +H +IG +G L K+ +Q V I F + +++ + G
Sbjct: 362 NSF---TVSSVAAPSWLHRFIIGKKGQNLAKITQQMPKVHIEF------TEGEDKITLEG 412
Query: 913 DKENVDKCCDILVEKANELL--YEWEERNQVREVKNSIKDRENVNSNNLKEK 1062
E+V+ + + +L+ ++ E N + + + N N +K++
Sbjct: 413 PTEDVNVAQEQIEGMVKDLINRMDYVEINIDHKFHRHLIGKSGANINRIKDQ 464
Score = 62.0 bits (149), Expect = 3e-09
Identities = 45/182 (24%), Positives = 80/182 (43%), Gaps = 10/182 (5%)
Frame = +1
Query: 412 ANKCLDFISIKGPRANLIDAK--------AGLDELIVVWENEKQDRIARNFENQVPVPIV 567
A CL+ + G L AK +G + ++ + R+ V +P
Sbjct: 101 AKICLEIMQRTGAHLELSLAKDQGLSIMVSGKLDAVMKARKDIVARLQTQASATVAIPKE 160
Query: 568 FHRKLIGQRGANIEMFRSKFNVSIQIPDQNSKSEEITIRGYEEDVKKAEDELLSMVDRWK 747
HR +IG+ G ++ K IQIP + S +I I G +E ++KA E+L +
Sbjct: 161 HHRFVIGKNGEKLQDLELKTATKIQIPRPDDPSNQIKITGTKEGIEKARHEVLLISAEQD 220
Query: 748 AQVIEELHIPRDVHPQLIGSRGSALRKLREQYNVEINFPSKNVDDDNANRVEVI--GDKE 921
+ +E L + + HP + G + ++ ++ IN P +V NR E++ G+KE
Sbjct: 221 KRAVERLEVEKAFHPFIAGPYNRLVGEIMQETGTRINIPPPSV-----NRTEIVFTGEKE 275
Query: 922 NV 927
+
Sbjct: 276 QL 277
>sp|P81021|VIGLN_CHICK Vigilin Length = 1270 Score = 203 bits (517), Expect = 7e-52 Identities = 132/393 (33%), Positives = 212/393 (53%), Gaps = 12/393 (3%) Frame = +1 Query: 4 QKHHRNLITRNMTEIKNIQNSFDVMIKFPNRDIS-SKSGENGYDEAHLNGNSDNTFVDTT 180 QK HR+++ + I+ I + V IKFP+R+ + + E E NG Sbjct: 883 QKFHRSIMGPKGSRIQQITRDYGVQIKFPDREENPAPVAEPALQE---NGEEGG------ 933 Query: 181 EEGDSNVEAEQNNP--ANWVALSGRRENCESAKEALLELVPVSEDVPLNNEFHRNFIGQK 354 EG +A+ ++P + + +SGRRE CE+AKEAL LVPV+ +V + + HR IGQK Sbjct: 934 -EGKDGKDADPSSPRKCDIIIISGRREKCEAAKEALQALVPVTIEVEVPFDLHRYIIGQK 992 Query: 355 GQGINTFSTKYNVVVNIP-PANKCLDFISIKGPRANLIDAKAGLDELIVVWENEKQDRIA 531 G GI ++ V + +P P + D I+I G ANL AKAGL E + + E++DR Sbjct: 993 GSGIRKMMDEFEVNIQVPAPELQSSDIITITGLAANLDRAKAGLLERVKELQAEQEDRAL 1052 Query: 532 RNFENQVPVPIVFHRKLIGQRGANIEMFRSKFNVSIQIPDQNSKSE---EITIRGYEEDV 702 R+F+ V V +H K+IG++GA I R++ V+IQ PD++ +S+ +ITI GYE++ Sbjct: 1053 RSFKLTVTVDPKYHPKIIGRKGAVITQIRTEHEVNIQFPDKDDESQAQDQITITGYEKNA 1112 Query: 703 KKAEDELLSMVDRWKAQVIEELHIPRDVHPQLIGSRGSALRKLREQYNVEINFPSKNVDD 882 + A D ++ +V + V E++ + VH ++IG+RG A+RK+ +++ V+I FP D Sbjct: 1113 EAARDAIMKIVGELEQMVSEDVTLDHRVHARIIGARGKAIRKIMDEFKVDIRFPQSGAPD 1172 Query: 883 DNANRVEVIGDKENVDKCCDILVEKANELLYEWEERNQVREVKNSIKDRENVNSNNLKEK 1062 N V V G ENV++ D ++ E L + + ++ E+ K Sbjct: 1173 PNC--VTVTGLPENVEEAIDHILNLEEEYLADVVDNEAMQVYMKPSSHEES------KVP 1224 Query: 1063 GPGFLLAGAPW-----QRVPDASSNVDFPTIDA 1146 GF++ AP ++ PD SS+ DFP+ A Sbjct: 1225 SKGFVVRDAPCGTVNNEKAPDMSSSEDFPSFGA 1257
Score = 103 bits (258), Expect = 8e-22
Identities = 77/300 (25%), Positives = 139/300 (46%), Gaps = 10/300 (3%)
Frame = +1
Query: 1 EQKHHRNLITRNMTEIKNIQNSFDVMIKFPNRDISSKSGENGYDEAHLNGNSDNTFVDTT 180
+ K HR+LI +N I I++ + V ++ P
Sbjct: 442 DHKFHRHLIGKNGANINRIKDLYKVSVRIP------------------------------ 471
Query: 181 EEGDSNVEAEQNNPANWVALSGRRENCESAKEALLELVPVSE-----DVPLNNEFHRNFI 345
N +N + + G + + AK+ LLEL E D+ + +FHR I
Sbjct: 472 ---------PDNEKSNLIRIEGDPQGVQQAKKELLELASRMENERTKDLIIEQKFHRTII 522
Query: 346 GQKGQGINTFSTKY-NVVVNIP-PANKCLDFISIKGPRANLIDAKAGLDELIVVWENEKQ 519
GQKG+ I K+ V++N P PA+K D + ++GP+ + + +++
Sbjct: 523 GQKGERIREIREKFPEVIINFPDPAHKS-DIVQLRGPKNEVEKCTKYMQKMVA------- 574
Query: 520 DRIARNFENQVPVPIVFHRKLIGQRGANIEMFRSKFNVSIQIPDQNSKSE-EITIRGYEE 696
D + +F VP+ FH+ +IG+ GANI+ R + N I +P + ++ ++ +G E+
Sbjct: 575 DLVENSFSISVPIFKQFHKNIIGKGGANIKKIREESNTKIDLPGREQATQRQLLSQGREQ 634
Query: 697 DVKKAEDELLSMVDRWKAQVIE-ELHIPRDVHPQLIGSRGSALRKLREQY-NVEINFPSK 870
VK +L+ + + A + E E+ IP +H LIG++G +R + E+ V I+FP++
Sbjct: 635 TVKLLRHRILA-IQKELANITEVEVSIPSKLHNSLIGAKGRFIRSIMEECGGVHIHFPTE 693
Score = 89.7 bits (221), Expect = 2e-17
Identities = 73/282 (25%), Positives = 133/282 (47%), Gaps = 7/282 (2%)
Frame = +1
Query: 148 GNSDNTFVDTTEEGDSNVEAEQNNPANWVALSGRRENCESAKEALLELVPVSEDVPLNNE 327
G N+ + E+ +VE + ++ + R E +AL E+ + +++
Sbjct: 311 GRKGNSLQEILEKTGVSVEIPPTDSSSETVIL--RGEPEKLGQALTEVYAKANSFTVSSV 368
Query: 328 -----FHRNFIGQKGQGINTFSTKY-NVVVNIPPANKCLDFISIKGPRANLIDAKAGLDE 489
HR IG GQ + + + + + D I+ +GP ++ A+ ++
Sbjct: 369 SAPSWLHRFIIGLFGQNLAKITQQMPKIHIEFTEGE---DKITSEGPTEDVNVAQEQIEV 425
Query: 490 LIVVWENEKQDRIARNFENQVPVPIVFHRKLIGQRGANIEMFRSKFNVSIQIPDQNSKSE 669
++ +D I R ++ V FHR LIG+ GANI + + VS++IP N KS
Sbjct: 426 MV-------KDLINRTDYAEINVDHKFHRHLIGKNGANINRIKDLYKVSVRIPPDNEKSN 478
Query: 670 EITIRGYEEDVKKAEDELLSMVDRWKAQVIEELHIPRDVHPQLIGSRGSALRKLREQY-N 846
I I G + V++A+ ELL + R + + ++L I + H +IG +G +R++RE++
Sbjct: 479 LIRIEGDPQGVQQAKKELLELASRMENERTKDLIIEQKFHRTIIGQKGERIREIREKFPE 538
Query: 847 VEINFPSKNVDDDNANRVEVIGDKENVDKCCDILVEKANELL 972
V INFP D V++ G K V+KC + + +L+
Sbjct: 539 VIINFPDPAHKSD---IVQLRGPKNEVEKCTKYMQKMVADLV 577
Score = 82.0 bits (201), Expect = 3e-15
Identities = 68/292 (23%), Positives = 135/292 (46%), Gaps = 39/292 (13%)
Frame = +1
Query: 232 VALSGRRENCESAKEALLELVP-----VSEDVPLNNEFHRNFIGQKGQGINTFSTKYN-V 393
+ + G E + A++ L L+ V + + ++ + HR+F+ ++GQ + + +Y V
Sbjct: 774 ITIMGTEEAVKEAQKELEALIKNLDNVVEDSMVVDPKHHRHFVIRRGQVLREIADEYGGV 833
Query: 394 VVNIPPANKCL-DFISIKGPRANLIDAKAGLDELIVVWENEKQDRIARNFENQVPVPIVF 570
+V +P + D +++KG + + AK + E+I + E Q I + +P F
Sbjct: 834 MVRLPTVSGTQSDKVTLKGAKDCVEAAKKRIQEIIE--DLEAQVTI------ECTIPQKF 885
Query: 571 HRKLIGQRGANIEMFRSKFNVSIQIPDQNS------------------------------ 660
HR ++G +G+ I+ + V I+ PD+
Sbjct: 886 HRSIMGPKGSRIQQITRDYGVQIKFPDREENPAPVAEPALQENGEEGGEGKDGKDADPSS 945
Query: 661 --KSEEITIRGYEEDVKKAEDELLSMVDRWKAQVIEELHIPRDVHPQLIGSRGSALRKLR 834
K + I I G E + A++ L ++V V E+ +P D+H +IG +GS +RK+
Sbjct: 946 PRKCDIIIISGRREKCEAAKEALQALV-----PVTIEVEVPFDLHRYIIGQKGSGIRKMM 1000
Query: 835 EQYNVEINFPSKNVDDDNANRVEVIGDKENVDKCCDILVEKANELLYEWEER 990
+++ V I P+ + +++ + + G N+D+ L+E+ EL E E+R
Sbjct: 1001 DEFEVNIQVPAPEL--QSSDIITITGLAANLDRAKAGLLERVKELQAEQEDR 1050
Score = 71.2 bits (173), Expect = 6e-12
Identities = 61/278 (21%), Positives = 123/278 (44%), Gaps = 4/278 (1%)
Frame = +1
Query: 49 KNIQNSFDVMIKFPNRDISSKSGENGYDEAHLNGNSDNTFVDTTEEGDSNVEAE-QNNPA 225
K + N +V + P++ +S G G + + EG + + P
Sbjct: 648 KELANITEVEVSIPSKLHNSLIGAKGRFIRSIMEECGGVHIHFPTEGSGSATVTIRAQPR 707
Query: 226 NWVALSGRRENCESAKEALLELVPVSEDVPLNNEFHRNFIGQKGQGINTFSTKYNVVVNI 405
W R C A+E + V D+ E+H+ IG+ G I +
Sbjct: 708 TW--RKPRSSCCTWAEEKQTKSYTV--DLRAKPEYHKFLIGKGGGNIRKVRDNTGARIIF 763
Query: 406 PPA-NKCLDFISIKGPRANLIDAKAGLDELIVVWENEKQDRIARNFENQVPVPIVFHRKL 582
P + +K + I+I G + +A+ L+ LI +N +D + V HR
Sbjct: 764 PTSEDKDQELITIMGTEEAVKEAQKELEALIKNLDNVVEDSMV--------VDPKHHRHF 815
Query: 583 IGQRGANIEMFRSKFN-VSIQIPD-QNSKSEEITIRGYEEDVKKAEDELLSMVDRWKAQV 756
+ +RG + ++ V +++P ++S+++T++G ++ V+ A+ + +++ +AQV
Sbjct: 816 VIRRGQVLREIADEYGGVMVRLPTVSGTQSDKVTLKGAKDCVEAAKKRIQEIIEDLEAQV 875
Query: 757 IEELHIPRDVHPQLIGSRGSALRKLREQYNVEINFPSK 870
E IP+ H ++G +GS ++++ Y V+I FP +
Sbjct: 876 TIECTIPQKFHRSIMGPKGSRIQQITRDYGVQIKFPDR 913
Score = 68.2 bits (165), Expect = 5e-11
Identities = 71/318 (22%), Positives = 122/318 (38%), Gaps = 9/318 (2%)
Frame = +1
Query: 1 EQKHHRNLITRNMTEIKNIQNSF-DVMIKFPNRDISSKSGENGYDEAHLNGNSDNTFVDT 177
EQK HR +I + I+ I+ F +V+I FP D AH
Sbjct: 514 EQKFHRTIIGQKGERIREIREKFPEVIINFP-------------DPAH------------ 548
Query: 178 TEEGDSNVEAEQNNPANWVALSGRRENCESAKEALLELVP------VSEDVPLNNEFHRN 339
++ V L G + E + + ++V S VP+ +FH+N
Sbjct: 549 --------------KSDIVQLRGPKNEVEKCTKYMQKMVADLVENSFSISVPIFKQFHKN 594
Query: 340 FIGQKGQGINTFSTKYNVVVNIPPANKCLDFISIKGPRANLIDAKAGLDELIVVWENEKQ 519
IG+ G I + N +++P + + R + K ++ + Q
Sbjct: 595 IIGKGGANIKKIREESNTKIDLPGREQATQRQLLSQGREQTV--KLLRHRILAI-----Q 647
Query: 520 DRIARNFENQVPVPIVFHRKLIGQRGANIEMFRSKFN-VSIQIPDQNSKSEEITIRGYEE 696
+A E +V +P H LIG +G I + V I P + S S +TIR
Sbjct: 648 KELANITEVEVSIPSKLHNSLIGAKGRFIRSIMEECGGVHIHFPTEGSGSATVTIRAQPR 707
Query: 697 DVKKAEDELLSMVDRWKAQVIE-ELHIPRDVHPQLIGSRGSALRKLREQYNVEINFPSKN 873
+K + + + + +L + H LIG G +RK+R+ I FP+
Sbjct: 708 TWRKPRSSCCTWAEEKQTKSYTVDLRAKPEYHKFLIGKGGGNIRKVRDNTGARIIFPTS- 766
Query: 874 VDDDNANRVEVIGDKENV 927
+D + + ++G +E V
Sbjct: 767 -EDKDQELITIMGTEEAV 783
Score = 65.1 bits (157), Expect = 4e-10
Identities = 47/180 (26%), Positives = 76/180 (42%), Gaps = 8/180 (4%)
Frame = +1
Query: 412 ANKCLDFISIKGPRANLIDAK--------AGLDELIVVWENEKQDRIARNFENQVPVPIV 567
A CLD + G L AK +G E ++ E R+ V +P
Sbjct: 100 AKICLDIMQKTGAHLELSLAKDQGLSIMVSGKLEAVMKARKEIVARLQTQASATVAIPKE 159
Query: 568 FHRKLIGQRGANIEMFRSKFNVSIQIPDQNSKSEEITIRGYEEDVKKAEDELLSMVDRWK 747
HR +IG+ G ++ K IQIP + S +I I G +E ++KA E+L +
Sbjct: 160 HHRFVIGKNGEKLQDLELKTATKIQIPRPDDPSNQIKITGTKEGIEKARHEILLISAEQD 219
Query: 748 AQVIEELHIPRDVHPQLIGSRGSALRKLREQYNVEINFPSKNVDDDNANRVEVIGDKENV 927
+ +E L + + HP + G + +L + IN P +V N + G+KE +
Sbjct: 220 KRAVERLDVEKVYHPFIAGPYNKLVSELMQDTGTRINIPPPSV---NKTEIVFTGEKEQL 276
Score = 64.3 bits (155), Expect = 7e-10
Identities = 60/291 (20%), Positives = 129/291 (44%), Gaps = 9/291 (3%)
Frame = +1
Query: 214 NNPANWVALSGRRENCESAKEALLELVPVSED------VPLNNEFHRNFIGQKGQGINTF 375
++P+N + ++G +E E A+ +L L+ +D + + +H G + ++
Sbjct: 189 DDPSNQIKITGTKEGIEKARHEIL-LISAEQDKRAVERLDVEKVYHPFIAGPYNKLVSEL 247
Query: 376 STKYNVVVNIPPANKCLDFISIKGPRANLIDAKAGLDELIVVWENEKQDRIARNFENQVP 555
+NIPP + I G + L A A + + ++E +K+ + V
Sbjct: 248 MQDTGTRINIPPPSVNKTEIVFTGEKEQLAQAVARVKK---IYEEKKK----KTTTIAVE 300
Query: 556 VPIVFHRKLIGQRGANIEMFRSKFNVSIQIPDQNSKSEEITIRGYEEDVKKAEDELLSMV 735
V H+ +IG++G +++ K VS++IP +S SE + +RG E + +A E+ +
Sbjct: 301 VKKSQHKYVIGRKGNSLQEILEKTGVSVEIPPTDSSSETVILRGEPEKLGQALTEVYAKA 360
Query: 736 DRWKAQVIEELHIPRDVHPQLIGSRGSALRKLREQY-NVEINFPSKNVDDDNANRVEVIG 912
+ + + + P +H +IG G L K+ +Q + I F + +++ G
Sbjct: 361 NSF---TVSSVSAPSWLHRFIIGLFGQNLAKITQQMPKIHIEF------TEGEDKITSEG 411
Query: 913 DKENVDKCCDILVEKANELL--YEWEERNQVREVKNSIKDRENVNSNNLKE 1059
E+V+ + + +L+ ++ E N + + + N N +K+
Sbjct: 412 PTEDVNVAQEQIEVMVKDLINRTDYAEINVDHKFHRHLIGKNGANINRIKD 462
>sp|Q8VDJ3|VIGLN_MOUSE Vigilin (High density lipoprotein-binding protein) (HDL-binding protein) Length = 1268 Score = 202 bits (515), Expect = 1e-51 Identities = 131/392 (33%), Positives = 209/392 (53%), Gaps = 11/392 (2%) Frame = +1 Query: 4 QKHHRNLITRNMTEIKNIQNSFDVMIKFPNRDISS-KSGENGYDEAHLNGNSDNTFVDTT 180 QK HR+++ + I+ I ++V IKFP+R+ + S E E NG D Sbjct: 882 QKFHRSVMGPKGSRIQQITRDYNVQIKFPDREENPVHSVEPSIQE---NG-------DEA 931 Query: 181 EEGDSNVEAEQNNP--ANWVALSGRRENCESAKEALLELVPVSEDVPLNNEFHRNFIGQK 354 EG E + +P + + +SGR+E CE+AKEAL LVPV+ +V + + HR IGQK Sbjct: 932 GEGREAKETDPGSPRRCDIIIISGRKEKCEAAKEALEALVPVTIEVEVPFDLHRYIIGQK 991 Query: 355 GQGINTFSTKYNVVVNIPPANKCLDFISIKGPRANLIDAKAGLDELIVVWENEKQDRIAR 534 G GI ++ V +++P D I+I G ANL AKAGL + + + E++DR R Sbjct: 992 GSGIRKMMDEFEVNIHVPAPELQSDTIAITGLAANLDRAKAGLLDRVKELQAEQEDRALR 1051 Query: 535 NFENQVPVPIVFHRKLIGQRGANIEMFRSKFNVSIQIPDQ---NSKSEEITIRGYEEDVK 705 +F+ V V +H K+IG++GA I R + V+IQ PD+ N ++ITI GYE++ + Sbjct: 1052 SFKLSVTVDPKYHPKIIGRKGAVITQIRLEHEVNIQFPDKDDGNQPQDQITITGYEKNTE 1111 Query: 706 KAEDELLSMVDRWKAQVIEELHIPRDVHPQLIGSRGSALRKLREQYNVEINFPSKNVDDD 885 A D +L +V + V E++ + VH ++IG+RG A+RK+ +++ V+I FP D Sbjct: 1112 AARDAILKIVGELEQMVSEDVPLDHRVHARIIGARGKAIRKIMDEFKVDIRFPQSGAPDP 1171 Query: 886 NANRVEVIGDKENVDKCCDILVEKANELLYEWEERNQVREVKNSIKDRENVNSNNLKEKG 1065 N V V G ENV++ D ++ E L + + ++ E+ + Sbjct: 1172 NC--VTVTGLPENVEEAIDHILNLEEEYLADVVDSEALQVYMKPPAHEES------RAPS 1223 Query: 1066 PGFLLAGAPW-----QRVPDASSNVDFPTIDA 1146 GF++ APW ++ PD SS+ +FP+ A Sbjct: 1224 KGFVVRDAPWTSNSSEKAPDMSSSEEFPSFGA 1255
Score = 114 bits (286), Expect = 5e-25
Identities = 84/330 (25%), Positives = 147/330 (44%), Gaps = 8/330 (2%)
Frame = +1
Query: 1 EQKHHRNLITRNMTEIKNIQNSFDVMIKFPNRDISSKSGENGYDEAHLNGNSDNTFVDTT 180
+ K HR+LI ++ I I++ + V ++ P
Sbjct: 443 DHKFHRHLIGKSGANINRIKDQYKVSVRIP------------------------------ 472
Query: 181 EEGDSNVEAEQNNPANWVALSGRRENCESAKEALLELVPVSE-----DVPLNNEFHRNFI 345
+ +N + + G + + AK LLEL E D+ + FHR I
Sbjct: 473 ---------PDSEKSNLIRIEGDPQGVQQAKRELLELASRMENERTKDLIIEQRFHRTII 523
Query: 346 GQKGQGINTFSTKY-NVVVNIPPANKCLDFISIKGPRANLIDAKAGLDELIVVWENEKQD 522
GQKG+ I K+ V++N P + D + ++GP+ + + +++ D
Sbjct: 524 GQKGERIREIRDKFPEVIINFPDPAQKSDIVQLRGPKNEVEKCTKYMQKMVA-------D 576
Query: 523 RIARNFENQVPVPIVFHRKLIGQRGANIEMFRSKFNVSIQIPDQNSKSEEITIRGYEEDV 702
+ ++ VP+ FH+ +IG+ GANI+ R + N I +P +NS SE I I G +
Sbjct: 577 LVENSYSISVPIFKQFHKNIIGKGGANIKKIREESNTKIDLPAENSNSETIIITGKRANC 636
Query: 703 KKAEDELLSMVDRWKAQVIE-ELHIPRDVHPQLIGSRGSALRKLREQY-NVEINFPSKNV 876
+ A +LS + + A + E E+ IP +H LIG++G +R + E+ V I+FP V
Sbjct: 637 EAARSRILS-IQKDLANIAEVEVSIPAKLHNSLIGTKGRLIRSIMEECGGVHIHFP---V 692
Query: 877 DDDNANRVEVIGDKENVDKCCDILVEKANE 966
+ ++ V + G +V+K L+ A E
Sbjct: 693 EGSGSDTVVIRGPSSDVEKAKKQLLHLAEE 722
Score = 103 bits (258), Expect = 8e-22
Identities = 85/305 (27%), Positives = 146/305 (47%), Gaps = 7/305 (2%)
Frame = +1
Query: 37 MTEIKNIQNSFDVM-IKFPNRDISSKSGENGYDEAHLNGNSDNTFVDTTEEGDSNVEAEQ 213
+TE+ NSF V + P+ G+ G + A + ++ TE G+ + E
Sbjct: 354 LTEVYAKANSFTVSSVSAPSWLHRFIIGKKGQNLAKITQQMPKVHIEFTE-GEDKITLE- 411
Query: 214 NNPANWVALSGRRENCESAKEALLELVPVSEDVPLNNEFHRNFIGQKGQGINTFSTKYNV 393
P V ++ +E E + L+ + E + ++++FHR+ IG+ G IN +Y V
Sbjct: 412 -GPTEDVNVA--QEQIEGMVKDLINRMDYVE-INIDHKFHRHLIGKSGANINRIKDQYKV 467
Query: 394 VVNIPPANKCLDFISIKGPRANLIDAKAGLDELIVVWENEKQDRIARNFENQVPVPIVFH 573
V IPP ++ + I I+G + AK L EL ENE+ + + FH
Sbjct: 468 SVRIPPDSEKSNLIRIEGDPQGVQQAKRELLELASRMENER--------TKDLIIEQRFH 519
Query: 574 RKLIGQRGANIEMFRSKF-NVSIQIPDQNSKSEEITIRGYEEDVKKAEDELLSMVDRWKA 750
R +IGQ+G I R KF V I PD KS+ + +RG + +V+K + MV A
Sbjct: 520 RTIIGQKGERIREIRDKFPEVIINFPDPAQKSDIVQLRGPKNEVEKCTKYMQKMV----A 575
Query: 751 QVIEELH-----IPRDVHPQLIGSRGSALRKLREQYNVEINFPSKNVDDDNANRVEVIGD 915
++E + I + H +IG G+ ++K+RE+ N +I+ P++N N+ + + G
Sbjct: 576 DLVENSYSISVPIFKQFHKNIIGKGGANIKKIREESNTKIDLPAEN---SNSETIIITGK 632
Query: 916 KENVD 930
+ N +
Sbjct: 633 RANCE 637
Score = 92.4 bits (228), Expect = 2e-18
Identities = 91/371 (24%), Positives = 166/371 (44%), Gaps = 43/371 (11%)
Frame = +1
Query: 1 EQKHHRNLITRNMTEIKNIQNSF-DVMIKFPN----RDISSKSG---------------- 117
EQ+ HR +I + I+ I++ F +V+I FP+ DI G
Sbjct: 515 EQRFHRTIIGQKGERIREIRDKFPEVIINFPDPAQKSDIVQLRGPKNEVEKCTKYMQKMV 574
Query: 118 ----ENGYD-------EAHLN--GNSDNTFVDTTEEGDSNVEAE-QNNPANWVALSGRRE 255
EN Y + H N G EE ++ ++ +N+ + + ++G+R
Sbjct: 575 ADLVENSYSISVPIFKQFHKNIIGKGGANIKKIREESNTKIDLPAENSNSETIIITGKRA 634
Query: 256 NCESAKEALL----ELVPVSE-DVPLNNEFHRNFIGQKGQGINTFSTKY-NVVVNIPPAN 417
NCE+A+ +L +L ++E +V + + H + IG KG+ I + + V ++ P
Sbjct: 635 NCEAARSRILSIQKDLANIAEVEVSIPAKLHNSLIGTKGRLIRSIMEECGGVHIHFPVEG 694
Query: 418 KCLDFISIKGPRANLIDAKAGLDELIVVWENEKQDRIARNFENQVPVPIVFHRKLIGQRG 597
D + I+GP +++ AK L L EKQ ++F + +H+ LIG+ G
Sbjct: 695 SGSDTVVIRGPSSDVEKAKKQLLHL----AEEKQ---TKSFTVDIRAKPEYHKFLIGKGG 747
Query: 598 ANIEMFRSKFNVSIQIPDQNSKSEE-ITIRGYEEDVKKAEDELLSMVDRWKAQVIEELHI 774
I R I P K ++ ITI G E+ V++A+ EL +++ + V + + +
Sbjct: 748 GKIRKVRDSTGARIIFPAAEDKDQDLITIIGKEDAVREAQKELEALIQNLENVVEDYMLV 807
Query: 775 PRDVHPQLIGSRGSALRKLREQY-NVEINFPSKNVDDDNANRVEVIGDKENVDKCCDILV 951
H + RG LR++ E+Y V ++FP D +V + G K+ C +
Sbjct: 808 DPKHHRHFVIRRGQVLREIAEEYGGVMVSFPRSGTQSD---KVTLKGAKD----CVEAAK 860
Query: 952 EKANELLYEWE 984
++ E++ + E
Sbjct: 861 KRIQEIIEDLE 871
Score = 91.3 bits (225), Expect = 5e-18
Identities = 62/216 (28%), Positives = 113/216 (52%), Gaps = 2/216 (0%)
Frame = +1
Query: 331 HRNFIGQKGQGINTFSTKY-NVVVNIPPANKCLDFISIKGPRANLIDAKAGLDELIVVWE 507
HR IG+KGQ + + + V + D I+++GP ++ A+ ++ ++
Sbjct: 376 HRFIIGKKGQNLAKITQQMPKVHIEFTEGE---DKITLEGPTEDVNVAQEQIEGMV---- 428
Query: 508 NEKQDRIARNFENQVPVPIVFHRKLIGQRGANIEMFRSKFNVSIQIPDQNSKSEEITIRG 687
+D I R ++ + FHR LIG+ GANI + ++ VS++IP + KS I I G
Sbjct: 429 ---KDLINRMDYVEINIDHKFHRHLIGKSGANINRIKDQYKVSVRIPPDSEKSNLIRIEG 485
Query: 688 YEEDVKKAEDELLSMVDRWKAQVIEELHIPRDVHPQLIGSRGSALRKLREQY-NVEINFP 864
+ V++A+ ELL + R + + ++L I + H +IG +G +R++R+++ V INFP
Sbjct: 486 DPQGVQQAKRELLELASRMENERTKDLIIEQRFHRTIIGQKGERIREIRDKFPEVIINFP 545
Query: 865 SKNVDDDNANRVEVIGDKENVDKCCDILVEKANELL 972
D V++ G K V+KC + + +L+
Sbjct: 546 DPAQKSD---IVQLRGPKNEVEKCTKYMQKMVADLV 578
Score = 81.3 bits (199), Expect = 6e-15
Identities = 57/228 (25%), Positives = 110/228 (48%), Gaps = 8/228 (3%)
Frame = +1
Query: 211 QNNPANWVALSGRRENCESAKEALLELVPVSE------DVPLNNEFHRNFIGQKGQGINT 372
+ + ++ V + G + E AK+ LL L + D+ E+H+ IG+ G I
Sbjct: 693 EGSGSDTVVIRGPSSDVEKAKKQLLHLAEEKQTKSFTVDIRAKPEYHKFLIGKGGGKIRK 752
Query: 373 FSTKYNVVVNIPPA-NKCLDFISIKGPRANLIDAKAGLDELIVVWENEKQDRIARNFENQ 549
+ P A +K D I+I G + +A+ L+ LI EN +D +
Sbjct: 753 VRDSTGARIIFPAAEDKDQDLITIIGKEDAVREAQKELEALIQNLENVVEDYML------ 806
Query: 550 VPVPIVFHRKLIGQRGANIEMFRSKFN-VSIQIPDQNSKSEEITIRGYEEDVKKAEDELL 726
V HR + +RG + ++ V + P ++S+++T++G ++ V+ A+ +
Sbjct: 807 --VDPKHHRHFVIRRGQVLREIAEEYGGVMVSFPRSGTQSDKVTLKGAKDCVEAAKKRIQ 864
Query: 727 SMVDRWKAQVIEELHIPRDVHPQLIGSRGSALRKLREQYNVEINFPSK 870
+++ +AQV E IP+ H ++G +GS ++++ YNV+I FP +
Sbjct: 865 EIIEDLEAQVTVECAIPQKFHRSVMGPKGSRIQQITRDYNVQIKFPDR 912
Score = 67.8 bits (164), Expect = 6e-11
Identities = 61/292 (20%), Positives = 131/292 (44%), Gaps = 9/292 (3%)
Frame = +1
Query: 214 NNPANWVALSGRRENCESAKEALLELVPVSED------VPLNNEFHRNFIGQKGQGINTF 375
++P+N + ++G +E E A+ +L L+ +D + + FH G + +
Sbjct: 190 DDPSNQIKITGTKEGIEKARHEVL-LISAEQDKRAVERLEVEKAFHPFIAGPYNRLVGEI 248
Query: 376 STKYNVVVNIPPANKCLDFISIKGPRANLIDAKAGLDELIVVWENEKQDRIARNFENQVP 555
+ +NIPP + I G + L A A + + ++E +K+ + V
Sbjct: 249 MQETGTRINIPPPSVNRTEIVFTGEKEQLAQAVARIKK---IYEEKKK----KTTTIAVE 301
Query: 556 VPIVFHRKLIGQRGANIEMFRSKFNVSIQIPDQNSKSEEITIRGYEEDVKKAEDELLSMV 735
V H+ +IG +G +++ + VS++IP +S SE + +RG E + +A E+ +
Sbjct: 302 VKKSQHKYVIGPKGNSLQEILERTGVSVEIPPSDSISETVILRGEPEKLGQALTEVYAKA 361
Query: 736 DRWKAQVIEELHIPRDVHPQLIGSRGSALRKLREQY-NVEINFPSKNVDDDNANRVEVIG 912
+ + + + P +H +IG +G L K+ +Q V I F + +++ + G
Sbjct: 362 NSF---TVSSVSAPSWLHRFIIGKKGQNLAKITQQMPKVHIEF------TEGEDKITLEG 412
Query: 913 DKENVDKCCDILVEKANELL--YEWEERNQVREVKNSIKDRENVNSNNLKEK 1062
E+V+ + + +L+ ++ E N + + + N N +K++
Sbjct: 413 PTEDVNVAQEQIEGMVKDLINRMDYVEINIDHKFHRHLIGKSGANINRIKDQ 464
Score = 65.1 bits (157), Expect = 4e-10
Identities = 46/182 (25%), Positives = 81/182 (44%), Gaps = 10/182 (5%)
Frame = +1
Query: 412 ANKCLDFISIKGPRANLIDAK--------AGLDELIVVWENEKQDRIARNFENQVPVPIV 567
A CL+ + G L AK +G + ++ + R+ VP+P
Sbjct: 101 AKICLEIMQRTGAHLELSLAKDQGLSIMVSGKLDAVMKARKDIVARLQTQASATVPIPKE 160
Query: 568 FHRKLIGQRGANIEMFRSKFNVSIQIPDQNSKSEEITIRGYEEDVKKAEDELLSMVDRWK 747
HR +IG+ G ++ K IQIP + S +I I G +E ++KA E+L +
Sbjct: 161 HHRFVIGKNGEKLQDLELKTATKIQIPRPDDPSNQIKITGTKEGIEKARHEVLLISAEQD 220
Query: 748 AQVIEELHIPRDVHPQLIGSRGSALRKLREQYNVEINFPSKNVDDDNANRVEVI--GDKE 921
+ +E L + + HP + G + ++ ++ IN P +V NR E++ G+KE
Sbjct: 221 KRAVERLEVEKAFHPFIAGPYNRLVGEIMQETGTRINIPPPSV-----NRTEIVFTGEKE 275
Query: 922 NV 927
+
Sbjct: 276 QL 277
Score = 56.6 bits (135), Expect = 1e-07
Identities = 51/247 (20%), Positives = 105/247 (42%), Gaps = 3/247 (1%)
Frame = +1
Query: 232 VALSGRRENCESAKEALLELVPV--SEDVPLNNEFHRNFIGQKGQGINTFSTKYNVVVNI 405
+ +SG+ + A++ ++ + S VP+ E HR IG+ G+ + K + I
Sbjct: 127 IMVSGKLDAVMKARKDIVARLQTQASATVPIPKEHHRFVIGKNGEKLQDLELKTATKIQI 186
Query: 406 PPANKCLDFISIKGPRANLIDAKAGLDELIVVWENEKQDRIARNFENQVPVPIVFHRKLI 585
P + + I I G + + KA + L++ E +K+ ++ V FH +
Sbjct: 187 PRPDDPSNQIKITGTKEGI--EKARHEVLLISAEQDKR------AVERLEVEKAFHPFIA 238
Query: 586 GQRGANIEMFRSKFNVSIQIPDQNSKSEEITIRGYEEDVKKAEDELLSMVDRWKAQVIE- 762
G + + I IP + EI G +E + +A + + + K +
Sbjct: 239 GPYNRLVGEIMQETGTRINIPPPSVNRTEIVFTGEKEQLAQAVARIKKIYEEKKKKTTTI 298
Query: 763 ELHIPRDVHPQLIGSRGSALRKLREQYNVEINFPSKNVDDDNANRVEVIGDKENVDKCCD 942
+ + + H +IG +G++L+++ E+ V + P D + V + G+ E + +
Sbjct: 299 AVEVKKSQHKYVIGPKGNSLQEILERTGVSVEIPP---SDSISETVILRGEPEKLGQALT 355
Query: 943 ILVEKAN 963
+ KAN
Sbjct: 356 EVYAKAN 362
>sp|Q9Z1A6|VIGLN_RAT Vigilin (High density lipoprotein-binding protein) (HDL-binding protein) Length = 1268 Score = 200 bits (508), Expect = 8e-51 Identities = 131/392 (33%), Positives = 207/392 (52%), Gaps = 11/392 (2%) Frame = +1 Query: 4 QKHHRNLITRNMTEIKNIQNSFDVMIKFPNRDISS-KSGENGYDEAHLNGNSDNTFVDTT 180 QK HR+++ + I+ I ++V IKFP+R+ + S E E NG D Sbjct: 882 QKFHRSVMGPKGSRIQQITRDYNVQIKFPDREENPVHSVEPSIQE---NG-------DEA 931 Query: 181 EEGDSNVEAEQNNP--ANWVALSGRRENCESAKEALLELVPVSEDVPLNNEFHRNFIGQK 354 EG E + +P + + +SGR+E CE+AKEAL LVPV+ +V + + HR IGQK Sbjct: 932 GEGREAKETDPGSPRRCDIIVISGRKEKCEAAKEALEALVPVTIEVEVPFDLHRYIIGQK 991 Query: 355 GQGINTFSTKYNVVVNIPPANKCLDFISIKGPRANLIDAKAGLDELIVVWENEKQDRIAR 534 G GI ++ V +++P I+I G ANL AKAGL + + + E++DR R Sbjct: 992 GSGIRKMMDEFEVNIHVPAPELQSHTIAITGLAANLDRAKAGLLDRVKELQAEQEDRALR 1051 Query: 535 NFENQVPVPIVFHRKLIGQRGANIEMFRSKFNVSIQIPDQ---NSKSEEITIRGYEEDVK 705 +F+ V V +H K+IG +GA I R + +V+IQ PD+ N ++ITI GYE++ + Sbjct: 1052 SFKLSVTVDPKYHPKIIGGKGAVITQIRLEHDVNIQFPDKDDGNQPQDQITITGYEKNTE 1111 Query: 706 KAEDELLSMVDRWKAQVIEELHIPRDVHPQLIGSRGSALRKLREQYNVEINFPSKNVDDD 885 A D +L +V + V E++ + VH ++IG+RG A+RK+ +++ V+I FP D Sbjct: 1112 AARDAILKIVGELEQMVSEDVPLDHRVHARIIGARGKAIRKIMDEFKVDIRFPQSGAPDP 1171 Query: 886 NANRVEVIGDKENVDKCCDILVEKANELLYEWEERNQVREVKNSIKDRENVNSNNLKEKG 1065 N V V G ENV++ D ++ E L + + ++ E+ K Sbjct: 1172 NC--VTVTGLPENVEEAIDHILNLEEEYLADVVDSEALQVYMKPPAHEES------KAPS 1223 Query: 1066 PGFLLAGAPW-----QRVPDASSNVDFPTIDA 1146 GF++ APW ++ PD SS+ + PT A Sbjct: 1224 KGFVVRDAPWTSNSSEKAPDMSSSEEIPTFGA 1255
Score = 113 bits (283), Expect = 1e-24
Identities = 83/330 (25%), Positives = 147/330 (44%), Gaps = 8/330 (2%)
Frame = +1
Query: 1 EQKHHRNLITRNMTEIKNIQNSFDVMIKFPNRDISSKSGENGYDEAHLNGNSDNTFVDTT 180
+ K HR+LI ++ I I++ + V ++ P
Sbjct: 443 DHKFHRHLIGKSGANINRIKDQYKVSVRIP------------------------------ 472
Query: 181 EEGDSNVEAEQNNPANWVALSGRRENCESAKEALLELVPVSE-----DVPLNNEFHRNFI 345
+ +N + + G + + AK LLEL E D+ + FHR I
Sbjct: 473 ---------PDSEKSNLIRIEGDPQGVQQAKRELLELASRMENERTKDLIIEQRFHRTII 523
Query: 346 GQKGQGINTFSTKY-NVVVNIPPANKCLDFISIKGPRANLIDAKAGLDELIVVWENEKQD 522
GQKG+ I K+ V++N P + + + ++GP+ + + +++ D
Sbjct: 524 GQKGERIREIRDKFPEVIINFPDPAQKSEIVQLRGPKNEVEKCTKYMQKMVA-------D 576
Query: 523 RIARNFENQVPVPIVFHRKLIGQRGANIEMFRSKFNVSIQIPDQNSKSEEITIRGYEEDV 702
+ ++ VP+ FH+ +IG+ GANI+ R + N I +P +NS SE I I G +
Sbjct: 577 LVENSYSISVPIFKQFHKNIIGKGGANIKKIREESNTKIDLPAENSNSETIVITGKRANC 636
Query: 703 KKAEDELLSMVDRWKAQVIE-ELHIPRDVHPQLIGSRGSALRKLREQY-NVEINFPSKNV 876
+ A +LS + + A + E E+ IP +H LIG++G +R + E+ V I+FP V
Sbjct: 637 EAARSRILS-IQKDLANIAEVEVSIPAKLHNSLIGTKGRLIRSIMEECGGVHIHFP---V 692
Query: 877 DDDNANRVEVIGDKENVDKCCDILVEKANE 966
+ ++ V + G +V+K L+ A E
Sbjct: 693 EGSGSDTVVIRGPSSDVEKAKKQLLHLAEE 722
Score = 105 bits (262), Expect = 3e-22
Identities = 86/305 (28%), Positives = 146/305 (47%), Gaps = 7/305 (2%)
Frame = +1
Query: 37 MTEIKNIQNSFDVM-IKFPNRDISSKSGENGYDEAHLNGNSDNTFVDTTEEGDSNVEAEQ 213
+TE+ NSF V + P+ G+ G + A + ++ TE G+ + E
Sbjct: 354 LTEVYAKANSFTVSSVSAPSWLHRFIIGKKGQNLAKITHQMPKVHIEFTE-GEDKITLE- 411
Query: 214 NNPANWVALSGRRENCESAKEALLELVPVSEDVPLNNEFHRNFIGQKGQGINTFSTKYNV 393
P V ++ +E E + L+ + E + ++++FHR+ IG+ G IN +Y V
Sbjct: 412 -GPTEDVNVA--QEQIEGMVKDLINRMDYVE-INIDHKFHRHLIGKSGANINRIKDQYKV 467
Query: 394 VVNIPPANKCLDFISIKGPRANLIDAKAGLDELIVVWENEKQDRIARNFENQVPVPIVFH 573
V IPP ++ + I I+G + AK L EL ENE+ + + FH
Sbjct: 468 SVRIPPDSEKSNLIRIEGDPQGVQQAKRELLELASRMENER--------TKDLIIEQRFH 519
Query: 574 RKLIGQRGANIEMFRSKF-NVSIQIPDQNSKSEEITIRGYEEDVKKAEDELLSMVDRWKA 750
R +IGQ+G I R KF V I PD KSE + +RG + +V+K + MV A
Sbjct: 520 RTIIGQKGERIREIRDKFPEVIINFPDPAQKSEIVQLRGPKNEVEKCTKYMQKMV----A 575
Query: 751 QVIEELH-----IPRDVHPQLIGSRGSALRKLREQYNVEINFPSKNVDDDNANRVEVIGD 915
++E + I + H +IG G+ ++K+RE+ N +I+ P++N N+ + + G
Sbjct: 576 DLVENSYSISVPIFKQFHKNIIGKGGANIKKIREESNTKIDLPAEN---SNSETIVITGK 632
Query: 916 KENVD 930
+ N +
Sbjct: 633 RANCE 637
Score = 91.3 bits (225), Expect = 5e-18
Identities = 63/219 (28%), Positives = 115/219 (52%), Gaps = 5/219 (2%)
Frame = +1
Query: 331 HRNFIGQKGQGINTFSTKY-NVVVNIPPANKCLDFISIKGPRANLIDAKAGLDELIVVWE 507
HR IG+KGQ + + + V + D I+++GP ++ A+ ++ ++
Sbjct: 376 HRFIIGKKGQNLAKITHQMPKVHIEFTEGE---DKITLEGPTEDVNVAQEQIEGMV---- 428
Query: 508 NEKQDRIARNFENQVPVPIVFHRKLIGQRGANIEMFRSKFNVSIQIPDQNSKSEEITIRG 687
+D I R ++ + FHR LIG+ GANI + ++ VS++IP + KS I I G
Sbjct: 429 ---KDLINRMDYVEINIDHKFHRHLIGKSGANINRIKDQYKVSVRIPPDSEKSNLIRIEG 485
Query: 688 YEEDVKKAEDELLSMVDRWKAQVIEELHIPRDVHPQLIGSRGSALRKLREQY-NVEINFP 864
+ V++A+ ELL + R + + ++L I + H +IG +G +R++R+++ V INFP
Sbjct: 486 DPQGVQQAKRELLELASRMENERTKDLIIEQRFHRTIIGQKGERIREIRDKFPEVIINFP 545
Query: 865 SKNVDDDNANRVEVI---GDKENVDKCCDILVEKANELL 972
D A + E++ G K V+KC + + +L+
Sbjct: 546 ------DPAQKSEIVQLRGPKNEVEKCTKYMQKMVADLV 578
Score = 90.1 bits (222), Expect = 1e-17
Identities = 90/371 (24%), Positives = 165/371 (44%), Gaps = 43/371 (11%)
Frame = +1
Query: 1 EQKHHRNLITRNMTEIKNIQNSF-DVMIKFPN----RDISSKSG---------------- 117
EQ+ HR +I + I+ I++ F +V+I FP+ +I G
Sbjct: 515 EQRFHRTIIGQKGERIREIRDKFPEVIINFPDPAQKSEIVQLRGPKNEVEKCTKYMQKMV 574
Query: 118 ----ENGYD-------EAHLN--GNSDNTFVDTTEEGDSNVEAE-QNNPANWVALSGRRE 255
EN Y + H N G EE ++ ++ +N+ + + ++G+R
Sbjct: 575 ADLVENSYSISVPIFKQFHKNIIGKGGANIKKIREESNTKIDLPAENSNSETIVITGKRA 634
Query: 256 NCESAKEALL----ELVPVSE-DVPLNNEFHRNFIGQKGQGINTFSTKY-NVVVNIPPAN 417
NCE+A+ +L +L ++E +V + + H + IG KG+ I + + V ++ P
Sbjct: 635 NCEAARSRILSIQKDLANIAEVEVSIPAKLHNSLIGTKGRLIRSIMEECGGVHIHFPVEG 694
Query: 418 KCLDFISIKGPRANLIDAKAGLDELIVVWENEKQDRIARNFENQVPVPIVFHRKLIGQRG 597
D + I+GP +++ AK L L EKQ ++F + +H+ LIG+ G
Sbjct: 695 SGSDTVVIRGPSSDVEKAKKQLLHL----AEEKQ---TKSFTVDIRAKPEYHKFLIGKGG 747
Query: 598 ANIEMFRSKFNVSIQIPDQNSKSEE-ITIRGYEEDVKKAEDELLSMVDRWKAQVIEELHI 774
I R I P K ++ ITI G E+ V++A+ EL +++ V + + +
Sbjct: 748 GKIRKVRDSTGARIIFPAAEDKDQDLITIIGKEDAVREAQKELEALIQNLDNVVEDYMLV 807
Query: 775 PRDVHPQLIGSRGSALRKLREQY-NVEINFPSKNVDDDNANRVEVIGDKENVDKCCDILV 951
H + RG LR++ E+Y V ++FP D +V + G K+ C +
Sbjct: 808 DPRHHRHFVIRRGQVLREIAEEYGGVMVSFPRSGTQSD---KVTLKGAKD----CVEAAK 860
Query: 952 EKANELLYEWE 984
++ E++ + E
Sbjct: 861 KRIQEIIEDLE 871
Score = 79.3 bits (194), Expect = 2e-14
Identities = 56/228 (24%), Positives = 110/228 (48%), Gaps = 8/228 (3%)
Frame = +1
Query: 211 QNNPANWVALSGRRENCESAKEALLELVPVSE------DVPLNNEFHRNFIGQKGQGINT 372
+ + ++ V + G + E AK+ LL L + D+ E+H+ IG+ G I
Sbjct: 693 EGSGSDTVVIRGPSSDVEKAKKQLLHLAEEKQTKSFTVDIRAKPEYHKFLIGKGGGKIRK 752
Query: 373 FSTKYNVVVNIPPA-NKCLDFISIKGPRANLIDAKAGLDELIVVWENEKQDRIARNFENQ 549
+ P A +K D I+I G + +A+ L+ LI +N +D +
Sbjct: 753 VRDSTGARIIFPAAEDKDQDLITIIGKEDAVREAQKELEALIQNLDNVVEDYML------ 806
Query: 550 VPVPIVFHRKLIGQRGANIEMFRSKFN-VSIQIPDQNSKSEEITIRGYEEDVKKAEDELL 726
V HR + +RG + ++ V + P ++S+++T++G ++ V+ A+ +
Sbjct: 807 --VDPRHHRHFVIRRGQVLREIAEEYGGVMVSFPRSGTQSDKVTLKGAKDCVEAAKKRIQ 864
Query: 727 SMVDRWKAQVIEELHIPRDVHPQLIGSRGSALRKLREQYNVEINFPSK 870
+++ +AQV E IP+ H ++G +GS ++++ YNV+I FP +
Sbjct: 865 EIIEDLEAQVTLECAIPQKFHRSVMGPKGSRIQQITRDYNVQIKFPDR 912
Score = 67.0 bits (162), Expect = 1e-10
Identities = 61/292 (20%), Positives = 130/292 (44%), Gaps = 9/292 (3%)
Frame = +1
Query: 214 NNPANWVALSGRRENCESAKEALLELVPVSED------VPLNNEFHRNFIGQKGQGINTF 375
++P+N + ++G +E E A+ +L L+ +D + + FH G + +
Sbjct: 190 DDPSNQIKITGTKEGIEKARHEVL-LISAEQDKRAVKRLEVEKAFHPFIAGPYNRLVGEI 248
Query: 376 STKYNVVVNIPPANKCLDFISIKGPRANLIDAKAGLDELIVVWENEKQDRIARNFENQVP 555
+ +NIPP + I G + L A A + + ++E +K+ + V
Sbjct: 249 MQETGTRINIPPPSVNRTEIVFTGEKEQLAQAVARIKK---IYEEKKK----KTTTIAVE 301
Query: 556 VPIVFHRKLIGQRGANIEMFRSKFNVSIQIPDQNSKSEEITIRGYEEDVKKAEDELLSMV 735
V H+ +IG +G +++ + VS++IP +S SE + +RG E + +A E+ +
Sbjct: 302 VKKSQHKYVIGPKGNSLQEILERTGVSVEIPPSDSISETVILRGEPEKLGQALTEVYAKA 361
Query: 736 DRWKAQVIEELHIPRDVHPQLIGSRGSALRKLREQY-NVEINFPSKNVDDDNANRVEVIG 912
+ + + + P +H +IG +G L K+ Q V I F + +++ + G
Sbjct: 362 NSF---TVSSVSAPSWLHRFIIGKKGQNLAKITHQMPKVHIEF------TEGEDKITLEG 412
Query: 913 DKENVDKCCDILVEKANELL--YEWEERNQVREVKNSIKDRENVNSNNLKEK 1062
E+V+ + + +L+ ++ E N + + + N N +K++
Sbjct: 413 PTEDVNVAQEQIEGMVKDLINRMDYVEINIDHKFHRHLIGKSGANINRIKDQ 464
Score = 63.5 bits (153), Expect = 1e-09
Identities = 45/182 (24%), Positives = 81/182 (44%), Gaps = 10/182 (5%)
Frame = +1
Query: 412 ANKCLDFISIKGPRANLIDAK--------AGLDELIVVWENEKQDRIARNFENQVPVPIV 567
A CL+ + G L AK +G + ++ + R+ VP+P
Sbjct: 101 AKICLEIMQRTGAHLELSLAKDQGLSIMVSGKLDAVMKARKDIVARLQTQASATVPIPKE 160
Query: 568 FHRKLIGQRGANIEMFRSKFNVSIQIPDQNSKSEEITIRGYEEDVKKAEDELLSMVDRWK 747
HR +IG+ G ++ K IQIP + S +I I G +E ++KA E+L +
Sbjct: 161 HHRFVIGKNGEKLQDLELKTATKIQIPRPDDPSNQIKITGTKEGIEKARHEVLLISAEQD 220
Query: 748 AQVIEELHIPRDVHPQLIGSRGSALRKLREQYNVEINFPSKNVDDDNANRVEVI--GDKE 921
+ ++ L + + HP + G + ++ ++ IN P +V NR E++ G+KE
Sbjct: 221 KRAVKRLEVEKAFHPFIAGPYNRLVGEIMQETGTRINIPPPSV-----NRTEIVFTGEKE 275
Query: 922 NV 927
+
Sbjct: 276 QL 277
Score = 57.4 bits (137), Expect = 9e-08
Identities = 53/247 (21%), Positives = 105/247 (42%), Gaps = 3/247 (1%)
Frame = +1
Query: 232 VALSGRRENCESAKEALLELVPV--SEDVPLNNEFHRNFIGQKGQGINTFSTKYNVVVNI 405
+ +SG+ + A++ ++ + S VP+ E HR IG+ G+ + K + I
Sbjct: 127 IMVSGKLDAVMKARKDIVARLQTQASATVPIPKEHHRFVIGKNGEKLQDLELKTATKIQI 186
Query: 406 PPANKCLDFISIKGPRANLIDAKAGLDELIVVWENEKQDRIARNFENQVPVPIVFHRKLI 585
P + + I I G + + KA + L++ E +K R + E V FH +
Sbjct: 187 PRPDDPSNQIKITGTKEGI--EKARHEVLLISAEQDK--RAVKRLE----VEKAFHPFIA 238
Query: 586 GQRGANIEMFRSKFNVSIQIPDQNSKSEEITIRGYEEDVKKAEDELLSMVDRWKAQVIE- 762
G + + I IP + EI G +E + +A + + + K +
Sbjct: 239 GPYNRLVGEIMQETGTRINIPPPSVNRTEIVFTGEKEQLAQAVARIKKIYEEKKKKTTTI 298
Query: 763 ELHIPRDVHPQLIGSRGSALRKLREQYNVEINFPSKNVDDDNANRVEVIGDKENVDKCCD 942
+ + + H +IG +G++L+++ E+ V + P D + V + G+ E + +
Sbjct: 299 AVEVKKSQHKYVIGPKGNSLQEILERTGVSVEIPP---SDSISETVILRGEPEKLGQALT 355
Query: 943 ILVEKAN 963
+ KAN
Sbjct: 356 EVYAKAN 362
>sp|P06105|SC160_YEAST Protein SCP160 (Protein HX) Length = 1222 Score = 79.3 bits (194), Expect = 2e-14 Identities = 79/325 (24%), Positives = 142/325 (43%), Gaps = 16/325 (4%) Frame = +1 Query: 7 KHHRNLITRNMTEIKNIQNSFDVMIKFP--NRDISSKSGENGYDEAHLNGNSDNTFVDTT 180 K H +LI + T +Q ++V I FP N ++ + G ++AH Sbjct: 722 KFHGSLIGPHGTYRNRLQEKYNVFINFPRDNEIVTIRGPSRGVNKAH------------- 768 Query: 181 EEGDSNVEAEQNNPANWVALSGRRENCESAKEALLELVPVSEDVPLNNEFHRNFIGQKGQ 360 EE + ++ E N V VP +E VP IG+ G Sbjct: 769 EELKALLDFEMENGHKMVIN-----------------VP-AEHVP-------RIIGKNGD 803 Query: 361 GINTFSTKYNVVVNI------PPANKCLDF-ISIKGPRANLIDAKAGLDELIVVWENEKQ 519 IN +Y V ++ P A + + + I G R N+ DA ++ ++ Sbjct: 804 NINDIRAEYGVEMDFLQKSTDPKAQETGEVELEITGSRQNIKDAAKRVESIVAE------ 857 Query: 520 DRIARNFENQV-PVPIVFHRKLIGQRGANIEMFRSKF------NVSIQIPDQNSKSEEIT 678 A +F +V + +H+ ++G G + SK N S+ IP+ +S++++IT Sbjct: 858 ---ASDFVTEVLKIDHKYHKSIVGSGGHILREIISKAGGEEIRNKSVDIPNADSENKDIT 914 Query: 679 IRGYEEDVKKAEDELLSMVDRWKAQVIEELHIPRDVHPQLIGSRGSALRKLREQYNVEIN 858 ++G ++ VKK +E+ +V + V + + IP + LIG G R+L ++N+ + Sbjct: 915 VQGPQKFVKKVVEEINKIVKDAENSVTKTIDIPAERKGALIGPGGIVRRQLESEFNINLF 974 Query: 859 FPSKNVDDDNANRVEVIGDKENVDK 933 P+K DD + ++ + G ENV+K Sbjct: 975 VPNK---DDPSGKITITGAPENVEK 996
Score = 75.1 bits (183), Expect = 4e-13
Identities = 83/319 (26%), Positives = 136/319 (42%), Gaps = 19/319 (5%)
Frame = +1
Query: 136 AHLNGNSDNTFVDTTEEGDSNVEA--EQNNPANW-----VALSGRRENCESAKEALLELV 294
A L GN + E+ ++ E+NN A+ V L+G N AK+ L
Sbjct: 647 ARLIGNKGSNLQQIREKFACQIDIPNEENNNASKDKTVEVTLTGLEYNLTHAKKYLAAEA 706
Query: 295 P-----VSEDVPLNNEFHRNFIGQKGQGINTFSTKYNVVVNIPPANKCLDFISIKGPRAN 459
+++++ + +FH + IG G N KYNV +N P N + ++I+GP
Sbjct: 707 KKWADIITKELIVPVKFHGSLIGPHGTYRNRLQEKYNVFINFPRDN---EIVTIRGPSRG 763
Query: 460 LIDAKAGLDELIVVWENEKQDRIARNFENQVPVPIVFHRKLIGQRGANIEMFRSKFNVSI 639
+ A L L+ +E E ++ N VP ++IG+ G NI R+++ V +
Sbjct: 764 VNKAHEELKALL-DFEMENGHKMVIN------VPAEHVPRIIGKNGDNINDIRAEYGVEM 816
Query: 640 QI-------PDQNSKSEEITIRGYEEDVKKAEDELLSMVDRWKAQVIEELHIPRDVHPQL 798
Q + E+ I G +++K A + S+V V E L I H +
Sbjct: 817 DFLQKSTDPKAQETGEVELEITGSRQNIKDAAKRVESIVAEASDFVTEVLKIDHKYHKSI 876
Query: 799 IGSRGSALRKLREQYNVEINFPSKNVDDDNANRVEVIGDKENVDKCCDILVEKANELLYE 978
+GS G LR++ + E +K+VD NA D EN DI V+ + +
Sbjct: 877 VGSGGHILREIISKAGGE-EIRNKSVDIPNA-------DSENK----DITVQGPQKFV-- 922
Query: 979 WEERNQVREVKNSIKDREN 1035
+ V E+ +KD EN
Sbjct: 923 ---KKVVEEINKIVKDAEN 938
Score = 73.2 bits (178), Expect = 2e-12
Identities = 39/114 (34%), Positives = 63/114 (55%), Gaps = 6/114 (5%)
Frame = +1
Query: 550 VPVPIVFHRKLIGQRGANIEMFRSKFNVSIQIPDQNS------KSEEITIRGYEEDVKKA 711
V +P +LIG +G+N++ R KF I IP++ + K+ E+T+ G E ++ A
Sbjct: 639 VNIPANSVARLIGNKGSNLQQIREKFACQIDIPNEENNNASKDKTVEVTLTGLEYNLTHA 698
Query: 712 EDELLSMVDRWKAQVIEELHIPRDVHPQLIGSRGSALRKLREQYNVEINFPSKN 873
+ L + +W + +EL +P H LIG G+ +L+E+YNV INFP N
Sbjct: 699 KKYLAAEAKKWADIITKELIVPVKFHGSLIGPHGTYRNRLQEKYNVFINFPRDN 752
Score = 45.1 bits (105), Expect = 4e-04
Identities = 53/246 (21%), Positives = 104/246 (42%), Gaps = 15/246 (6%)
Frame = +1
Query: 190 DSNVEAEQNNPANWVALSGRRENCESAKEALLELV--PVSEDVPLNNEFHRNFIGQKGQG 363
D +VE+ + A +SG N AK L++ + P++ + + ++ + IG G+
Sbjct: 140 DVSVESTLSKNARTFLVSGVAANVHEAKRELVKKLTKPINAVIEVPSKCKASIIGSGGRT 199
Query: 364 INTFSTKYNVVVNIPPA----------NKCLDFISIKGPRANLIDAKAGLDELIVVWENE 513
I S Y V +N+ + +S+ G ++ AKA + L +V E
Sbjct: 200 IREISDAYEVKINVSKEVNENSYDEDMDDTTSNVSLFGDFESVNLAKAKI--LAIVKEET 257
Query: 514 KQDRIARNFENQVPVPIVFHRKLIGQRG---ANIEMFRSKFNVSIQIPDQNSKSEEITIR 684
K I E++ +P + + G ++ ++ ++ I P + +K+ + +I+
Sbjct: 258 KNATIKLVVEDEKYLPYIDVSEFASDEGDEEVKVQFYKKSGDIVILGpreKAKATKTSIQ 317
Query: 685 GYEEDVKKAEDELLSMVDRWKAQVIEELHIPRDVHPQLIGSRGSALRKLREQYNVEINFP 864
Y +KK L S +D K ++ + D +L+E+YNV + FP
Sbjct: 318 DY---LKK----LASNLDEEKVKIPSKFQFLIDAE------------ELKEKYNVIVTFP 358
Query: 865 SKNVDD 882
S D+
Sbjct: 359 STPDDE 364
Score = 43.9 bits (102), Expect = 0.001
Identities = 24/91 (26%), Positives = 55/91 (60%)
Frame = +1
Query: 637 IQIPDQNSKSEEITIRGYEEDVKKAEDELLSMVDRWKAQVIEELHIPRDVHPQLIGSRGS 816
+QI + ++T+RG E+ K A S+++ ++ ++IP + +LIG++GS
Sbjct: 596 LQIKLHTPEENQLTVRGDEKAAKAANKIFESILNSPSSKSKMTVNIPANSVARLIGNKGS 655
Query: 817 ALRKLREQYNVEINFPSKNVDDDNANRVEVI 909
L+++RE++ +I+ P N +++NA++ + +
Sbjct: 656 NLQQIREKFACQIDIP--NEENNNASKDKTV 684
Score = 31.2 bits (69), Expect = 6.6
Identities = 19/54 (35%), Positives = 32/54 (59%), Gaps = 1/54 (1%)
Frame = +1
Query: 796 LIGSRGSALRKLREQYNVEINFPSKNVDDDNANRVEVI-GDKENVDKCCDILVE 954
++G GS ++K+RE +V IN P K+ D N V I G K V+K +++++
Sbjct: 1168 IVGPGGSNIKKIREAADVIINVPRKS---DKVNDVVYIRGTKAGVEKAGEMVLK 1218
>sp|Q80VL1|TDRKH_MOUSE Tudor and KH domain-containing protein Length = 560 Score = 42.7 bits (99), Expect = 0.002 Identities = 41/215 (19%), Positives = 97/215 (45%), Gaps = 6/215 (2%) Frame = +1 Query: 535 NFENQVPVPIVFHRKLIGQRGANIEMFRSKFNVSIQIPDQNSKSEEI-TIRGYEEDVKKA 711 + E ++ VP + +IG++GANI+ R + I + ++ E + I G+ V KA Sbjct: 52 DIEIEMRVPQEAVKLIIGRQGANIKQLRKQTGARIDVDTEDVGDERVLLISGFPVQVCKA 111 Query: 712 EDELLSMVDRWKAQVIEELHIPRDVHPQLIGSRGSALRKLREQYNVEINFPSKNVDDDNA 891 + + ++ V E+L +P+ ++IG G +R + + +I ++ Sbjct: 112 KAAIHQILTE-NTPVFEQLSVPQRSVGRIIGRGGETIRSICKASGAKITCDKESEGTLLL 170 Query: 892 NR-VEVIGDKENVDKCCDILVEKANELLYEWEERNQV-REVKNSIKDRENVNSNNLKEKG 1065 +R +++ G ++ V +++EK +E + E R ++ + + ++ ++ + Sbjct: 171 SRLIKISGTQKEVAAAKHLILEKVSE---DEELRKRIAHSAETRVPRKQPISVRREEVTE 227 Query: 1066 PGFLLAGAPWQRVPDA---SSNVDFPTIDANGDKI 1161 PG A W+ + ++ ++ P GD + Sbjct: 228 PGGAGEAALWKNTNSSMGPATPLEVPLRKGGGDMV 262
>sp|Q9Y2W6|TDRKH_HUMAN Tudor and KH domain-containing protein Length = 606 Score = 41.6 bits (96), Expect = 0.005 Identities = 32/146 (21%), Positives = 71/146 (48%), Gaps = 2/146 (1%) Frame = +1 Query: 535 NFENQVPVPIVFHRKLIGQRGANIEMFRSKFNVSIQIPDQNSKSEEI-TIRGYEEDVKKA 711 + E ++ VP + +IG++GANI+ R + I + ++ E + I G+ V KA Sbjct: 52 DIEIEMRVPQEAVKLIIGRQGANIKQLRKQTGARIDVDTEDVGDERVLLISGFPVQVCKA 111 Query: 712 EDELLSMVDRWKAQVIEELHIPRDVHPQLIGSRGSALRKLREQYNVEINFPSKNVDDDNA 891 + + ++ V E+L +P+ ++IG G +R + + +I ++ Sbjct: 112 KAAIHQILTE-NTPVSEQLSVPQRSVGRIIGRGGETIRSICKASGAKITCDKESEGTLLL 170 Query: 892 NR-VEVIGDKENVDKCCDILVEKANE 966 +R +++ G ++ V +++EK +E Sbjct: 171 SRLIKISGTQKEVAAAKHLILEKVSE 196
>sp|P57722|PCBP3_MOUSE Poly(rC)-binding protein 3 (Alpha-CP3) Length = 339 Score = 40.8 bits (94), Expect = 0.008 Identities = 32/141 (22%), Positives = 60/141 (42%), Gaps = 16/141 (11%) Frame = +1 Query: 580 LIGQRGANIEMFRSKFNVSIQIPDQNSKSEEITIRGYEEDVKKA---------EDELLSM 732 +IG++G ++ R + I I + N +TI G + + KA ED + SM Sbjct: 28 IIGKKGETVKKMREESGARINISEGNCPERIVTITGPTDAIFKAFAMIAYKFEEDIINSM 87 Query: 733 VDR---WKAQVIEELHIPRDVHPQLIGSRGSALRKLREQYNVEINFPSKNVDDDNANRVE 903 + K V L +P LIG GS ++++RE ++ + + V Sbjct: 88 SNSPATSKPPVTLRLVVPASQCGSLIGKGGSKIKEIRESTGAQVQVAGDMLPNSTERAVT 147 Query: 904 VIGDKENVDKC----CDILVE 954 + G + + +C C +++E Sbjct: 148 ISGTPDAIIQCVKQICVVMLE 168
>sp|P57721|PCBP3_HUMAN Poly(rC)-binding protein 3 (Alpha-CP3) Length = 339 Score = 40.8 bits (94), Expect = 0.008 Identities = 32/141 (22%), Positives = 60/141 (42%), Gaps = 16/141 (11%) Frame = +1 Query: 580 LIGQRGANIEMFRSKFNVSIQIPDQNSKSEEITIRGYEEDVKKA---------EDELLSM 732 +IG++G ++ R + I I + N +TI G + + KA ED + SM Sbjct: 28 IIGKKGETVKKMREESGARINISEGNCPERIVTITGPTDAIFKAFAMIAYKFEEDIINSM 87 Query: 733 VDR---WKAQVIEELHIPRDVHPQLIGSRGSALRKLREQYNVEINFPSKNVDDDNANRVE 903 + K V L +P LIG GS ++++RE ++ + + V Sbjct: 88 SNSPATSKPPVTLRLVVPASQCGSLIGKGGSKIKEIRESTGAQVQVAGDMLPNSTERAVT 147 Query: 904 VIGDKENVDKC----CDILVE 954 + G + + +C C +++E Sbjct: 148 ISGTPDAIIQCVKQICVVMLE 168
>sp|Q01570|SECA_PAVLU Preprotein translocase secA subunit Length = 891 Score = 38.9 bits (89), Expect = 0.031 Identities = 50/186 (26%), Positives = 82/186 (44%), Gaps = 20/186 (10%) Frame = +1 Query: 565 VFHRKLIGQRGANI---EMFRSKFNVSIQIPDQNSKSEEITIRGYEEDVKKAEDELLSMV 735 +F++KL G G + + F+ +N+S+ N K I +EDV S+ Sbjct: 357 LFYKKLSGMTGTALTEAKEFKKIYNLSVDCVPINKKVNRID----KEDVVYK-----SLY 407 Query: 736 DRWKAQVIEELHIPRDVHPQLIGS----RGSALRKLREQYNVEINFPSKNVDDDNANRVE 903 +WKA + E L I P LIG+ + L ++YN++ + + ++ AN E Sbjct: 408 AKWKAVLYESLSIHEQGRPLLIGTSNVKNSEIVSGLLKEYNIKHSLLNAK-PENAANESE 466 Query: 904 VI---GDKENV-------DKCCDILVEKANELLYEWEERNQVREVKNSIKDR---ENVNS 1044 +I G K +V + DIL+ + L + E R R + S+ D +N Sbjct: 467 IIAQAGRKGSVTIATNMAGRGTDILLGGNPDFLTKGELRYIFRSIVLSLDDMTVPKNELI 526 Query: 1045 NNLKEK 1062 NNLK K Sbjct: 527 NNLKYK 532
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.317 0.136 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 137,401,078
Number of Sequences: 369166
Number of extensions: 2815626
Number of successful extensions: 9395
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 8678
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 9262
length of database: 68,354,980
effective HSP length: 113
effective length of database: 47,479,925
effective search space used: 14433897200
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail