DrC_00456
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
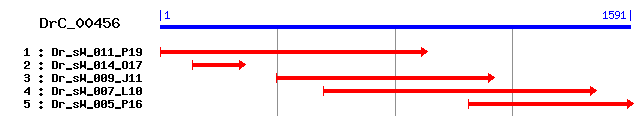
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00456 (1591 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P21573|YBOX1_XENLA Nuclease sensitive element binding pr... 122 2e-27 sp|Q9JKB3|DBPA_MOUSE DNA-binding protein A (Cold shock doma... 122 3e-27 sp|P16989|DBPA_HUMAN DNA-binding protein A (Cold shock doma... 122 3e-27 sp|Q62764|DBPA_RAT DNA-binding protein A (Cold shock domain... 122 3e-27 sp|P62960|YBOX1_MOUSE Nuclease sensitive element binding pr... 121 5e-27 sp|P67809|YBOX1_HUMAN Nuclease sensitive element binding pr... 121 5e-27 sp|Q06066|YBOX1_CHICK Nuclease sensitive element binding pr... 121 5e-27 sp|P41824|YBOXH_APLCA Y-box factor homolog (APY1) 117 1e-25 sp|Q00436|YB3_XENLA B box binding protein (YB3 protein) 116 2e-25 sp|P21574|YBX2A_XENLA Y-box binding protein 2-A (Cytoplasmi... 110 9e-24
>sp|P21573|YBOX1_XENLA Nuclease sensitive element binding protein 1 (Y-box binding protein 1) (Y-box transcription factor) Length = 303 Score = 122 bits (307), Expect = 2e-27 Identities = 57/91 (62%), Positives = 67/91 (73%) Frame = +1 Query: 91 LATKINGKVKWFNVKSGYGFINRNDNQEDIFVHQTAILKNNPRKWQRSXXXXXXXXXXXX 270 +ATK+ G VKWFNV++GYGFINRND +ED+FVHQTAI KNNPRK+ RS Sbjct: 33 IATKVLGTVKWFNVRNGYGFINRNDTKEDVFVHQTAIKKNNPRKYLRSVGDGETVEFDVV 92 Query: 271 XXXXXLEAANVTGPDGIPVQGSKYAADKRHY 363 EAANVTGP+G+PVQGSKYAAD+ HY Sbjct: 93 EGEKGAEAANVTGPEGVPVQGSKYAADRNHY 123
>sp|Q9JKB3|DBPA_MOUSE DNA-binding protein A (Cold shock domain protein A) (Y-box protein 3) sp|Q5PQD5|DBPA_XENLA DNA-binding protein A (Cold shock domain protein A) Length = 361 Score = 122 bits (306), Expect = 3e-27 Identities = 60/100 (60%), Positives = 71/100 (71%) Frame = +1 Query: 64 EKKILAYDYLATKINGKVKWFNVKSGYGFINRNDNQEDIFVHQTAILKNNPRKWQRSXXX 243 EKK+LA TK+ G VKWFNV++GYGFINRND +ED+FVHQTAI KNNPRK+ RS Sbjct: 75 EKKVLA-----TKVLGTVKWFNVRNGYGFINRNDTKEDVFVHQTAIKKNNPRKYLRSVGD 129 Query: 244 XXXXXXXXXXXXXXLEAANVTGPDGIPVQGSKYAADKRHY 363 EAANVTGPDG+PV+GS+YAAD+R Y Sbjct: 130 GETVEFDVVEGEKGAEAANVTGPDGVPVEGSRYAADRRRY 169
>sp|P16989|DBPA_HUMAN DNA-binding protein A (Cold shock domain protein A) (Single-strand DNA binding protein NF-GMB) Length = 372 Score = 122 bits (306), Expect = 3e-27 Identities = 60/100 (60%), Positives = 71/100 (71%) Frame = +1 Query: 64 EKKILAYDYLATKINGKVKWFNVKSGYGFINRNDNQEDIFVHQTAILKNNPRKWQRSXXX 243 EKK+LA TK+ G VKWFNV++GYGFINRND +ED+FVHQTAI KNNPRK+ RS Sbjct: 83 EKKVLA-----TKVLGTVKWFNVRNGYGFINRNDTKEDVFVHQTAIKKNNPRKYLRSVGD 137 Query: 244 XXXXXXXXXXXXXXLEAANVTGPDGIPVQGSKYAADKRHY 363 EAANVTGPDG+PV+GS+YAAD+R Y Sbjct: 138 GETVEFDVVEGEKGAEAANVTGPDGVPVEGSRYAADRRRY 177
>sp|Q62764|DBPA_RAT DNA-binding protein A (Cold shock domain protein A) (Muscle Y-box protein YB2) (Y-box binding protein-A) (RYB-A) Length = 361 Score = 122 bits (306), Expect = 3e-27 Identities = 60/100 (60%), Positives = 71/100 (71%) Frame = +1 Query: 64 EKKILAYDYLATKINGKVKWFNVKSGYGFINRNDNQEDIFVHQTAILKNNPRKWQRSXXX 243 EKK+LA TK+ G VKWFNV++GYGFINRND +ED+FVHQTAI KNNPRK+ RS Sbjct: 75 EKKVLA-----TKVLGTVKWFNVRNGYGFINRNDTKEDVFVHQTAIKKNNPRKYLRSVGD 129 Query: 244 XXXXXXXXXXXXXXLEAANVTGPDGIPVQGSKYAADKRHY 363 EAANVTGPDG+PV+GS+YAAD+R Y Sbjct: 130 GETVEFDVVEGEKGAEAANVTGPDGVPVEGSRYAADRRRY 169
>sp|P62960|YBOX1_MOUSE Nuclease sensitive element binding protein 1 (Y-box binding protein 1) (Y-box transcription factor) (YB-1) (CCAAT-binding transcription factor I subunit A) (CBF-A) (Enhancer factor I subunit A) (EFI-A) (DNA-binding protein B) (DBPB) sp|P62961|YBOX1_RAT Nuclease sensitive element binding protein 1 (Y-box binding protein 1) (Y-box transcription factor) (YB-1) (CCAAT-binding transcription factor I subunit A) (CBF-A) (Enhancer factor I subunit A) (EFI-A) (DNA-binding protein B) (DBPB) Length = 322 Score = 121 bits (304), Expect = 5e-27 Identities = 57/91 (62%), Positives = 66/91 (72%) Frame = +1 Query: 91 LATKINGKVKWFNVKSGYGFINRNDNQEDIFVHQTAILKNNPRKWQRSXXXXXXXXXXXX 270 +ATK+ G VKWFNV++GYGFINRND +ED+FVHQTAI KNNPRK+ RS Sbjct: 53 IATKVLGTVKWFNVRNGYGFINRNDTKEDVFVHQTAIKKNNPRKYLRSVGDGETVEFDVV 112 Query: 271 XXXXXLEAANVTGPDGIPVQGSKYAADKRHY 363 EAANVTGP G+PVQGSKYAAD+ HY Sbjct: 113 EGEKGAEAANVTGPGGVPVQGSKYAADRNHY 143
>sp|P67809|YBOX1_HUMAN Nuclease sensitive element binding protein 1 (Y-box binding protein 1) (Y-box transcription factor) (YB-1) (CCAAT-binding transcription factor I subunit A) (CBF-A) (Enhancer factor I subunit A) (EFI-A) (DNA-binding protein B) (DBPB) sp|P67808|YBOX1_BOVIN Nuclease sensitive element binding protein 1 (Y-box binding protein 1) (Y-box transcription factor) (YB-1) (CCAAT-binding transcription factor I subunit A) (CBF-A) (Enhancer factor I subunit A) (EFI-A) (DNA-binding protein B) (DBPB) Length = 324 Score = 121 bits (304), Expect = 5e-27 Identities = 57/91 (62%), Positives = 66/91 (72%) Frame = +1 Query: 91 LATKINGKVKWFNVKSGYGFINRNDNQEDIFVHQTAILKNNPRKWQRSXXXXXXXXXXXX 270 +ATK+ G VKWFNV++GYGFINRND +ED+FVHQTAI KNNPRK+ RS Sbjct: 55 IATKVLGTVKWFNVRNGYGFINRNDTKEDVFVHQTAIKKNNPRKYLRSVGDGETVEFDVV 114 Query: 271 XXXXXLEAANVTGPDGIPVQGSKYAADKRHY 363 EAANVTGP G+PVQGSKYAAD+ HY Sbjct: 115 EGEKGAEAANVTGPGGVPVQGSKYAADRNHY 145
>sp|Q06066|YBOX1_CHICK Nuclease sensitive element binding protein 1 (Y-box binding protein 1) (Y-box transcription factor) Length = 321 Score = 121 bits (304), Expect = 5e-27 Identities = 57/91 (62%), Positives = 66/91 (72%) Frame = +1 Query: 91 LATKINGKVKWFNVKSGYGFINRNDNQEDIFVHQTAILKNNPRKWQRSXXXXXXXXXXXX 270 +ATK+ G VKWFNV++GYGFINRND +ED+FVHQTAI KNNPRK+ RS Sbjct: 52 IATKVLGTVKWFNVRNGYGFINRNDTKEDVFVHQTAIKKNNPRKYLRSVGDGETVEFDVV 111 Query: 271 XXXXXLEAANVTGPDGIPVQGSKYAADKRHY 363 EAANVTGP G+PVQGSKYAAD+ HY Sbjct: 112 EGEKGAEAANVTGPGGVPVQGSKYAADRNHY 142
>sp|P41824|YBOXH_APLCA Y-box factor homolog (APY1) Length = 253 Score = 117 bits (292), Expect = 1e-25 Identities = 63/124 (50%), Positives = 79/124 (63%), Gaps = 8/124 (6%) Frame = +1 Query: 16 MADIQTSPT--------ERSVHATEKKILAYDYLATKINGKVKWFNVKSGYGFINRNDNQ 171 MAD + P E++ EKKI+A ++++G VKWFNVKSGYGFINR+D + Sbjct: 1 MADTEKQPEVEENQPDQEQNEEQKEKKIIA-----SQVSGTVKWFNVKSGYGFINRDDTK 55 Query: 172 EDIFVHQTAILKNNPRKWQRSXXXXXXXXXXXXXXXXXLEAANVTGPDGIPVQGSKYAAD 351 ED+FVHQTAI+KNNPRK+ RS EAANVTGP+G VQGSKYAAD Sbjct: 56 EDVFVHQTAIVKNNPRKYLRSVGDGEKVEFDVVEGEKGNEAANVTGPEGSNVQGSKYAAD 115 Query: 352 KRHY 363 +R + Sbjct: 116 RRRF 119
>sp|Q00436|YB3_XENLA B box binding protein (YB3 protein) Length = 305 Score = 116 bits (291), Expect = 2e-25 Identities = 58/100 (58%), Positives = 69/100 (69%) Frame = +1 Query: 64 EKKILAYDYLATKINGKVKWFNVKSGYGFINRNDNQEDIFVHQTAILKNNPRKWQRSXXX 243 EKK++A TK+ G VKWFNV++GYGFINRND +ED+FVHQTAI KNNPRK+ RS Sbjct: 29 EKKVIA-----TKVLGTVKWFNVRNGYGFINRNDTKEDVFVHQTAIKKNNPRKYLRSVGD 83 Query: 244 XXXXXXXXXXXXXXLEAANVTGPDGIPVQGSKYAADKRHY 363 EAANVTGP +PVQGSKYAAD+ +Y Sbjct: 84 GETVEFDVVEGEKGAEAANVTGPGPVPVQGSKYAADRNNY 123
>sp|P21574|YBX2A_XENLA Y-box binding protein 2-A (Cytoplasmic RNA-binding protein p56) (mRNP4) Length = 336 Score = 110 bits (276), Expect = 9e-24 Identities = 51/91 (56%), Positives = 65/91 (71%) Frame = +1 Query: 91 LATKINGKVKWFNVKSGYGFINRNDNQEDIFVHQTAILKNNPRKWQRSXXXXXXXXXXXX 270 LAT++ G VKWFNV++GYGFINRND +ED+FVHQTAI KNNPRK+ RS Sbjct: 38 LATQVQGTVKWFNVRNGYGFINRNDTKEDVFVHQTAIKKNNPRKFLRSVGDGETVEFDVV 97 Query: 271 XXXXXLEAANVTGPDGIPVQGSKYAADKRHY 363 EAANVTGP G+PV+GS++A ++R + Sbjct: 98 EGEKGAEAANVTGPGGVPVKGSRFAPNRRRF 128
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 148,990,278
Number of Sequences: 369166
Number of extensions: 2565469
Number of successful extensions: 5778
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 5549
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5755
length of database: 68,354,980
effective HSP length: 115
effective length of database: 47,110,455
effective search space used: 19503728370
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail