DrC_00357
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
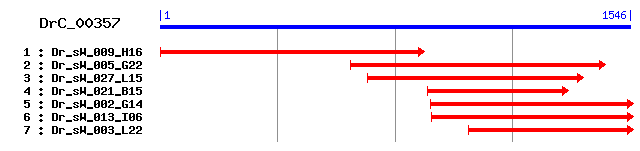
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00357 (1546 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q61879|MYH10_MOUSE Myosin-10 (Myosin heavy chain, nonmus... 241 5e-63 sp|Q9JLT0|MYH10_RAT Myosin-10 (Myosin heavy chain, nonmuscl... 241 5e-63 sp|Q27991|MYH10_BOVIN Myosin-10 (Myosin heavy chain, nonmus... 241 5e-63 sp|P35580|MYH10_HUMAN Myosin-10 (Myosin heavy chain, nonmus... 238 3e-62 sp|P35579|MYH9_HUMAN Myosin-9 (Myosin heavy chain, nonmuscl... 238 4e-62 sp|Q8VDD5|MYH9_MOUSE Myosin-9 (Myosin heavy chain, nonmuscl... 238 4e-62 sp|Q99105|MYSU_RABIT Myosin heavy chain, embryonic smooth m... 238 4e-62 sp|P14105|MYH9_CHICK Myosin-9 (Myosin heavy chain, nonmuscl... 235 3e-61 sp|Q62812|MYH9_RAT Myosin-9 (Myosin heavy chain, nonmuscle ... 233 1e-60 sp|P35749|MYH11_HUMAN Myosin-11 (Myosin heavy chain, smooth... 232 3e-60
>sp|Q61879|MYH10_MOUSE Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B) Length = 1976 Score = 241 bits (614), Expect = 5e-63 Identities = 138/422 (32%), Positives = 227/422 (53%) Frame = +3 Query: 18 LNRQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKERIEMQ 197 L + +L+S+KDDVGKN+ L+ +K LE +E + +EELE+E + K R+E+ Sbjct: 1509 LRADMEDLMSSKDDVGKNVHELEKSKRALEQQVEEMRTQLEELEDELQATEDAKLRLEVN 1568 Query: 198 LTATKAQLDRELSSKDEMHDEQFKILNKKLRDTEATLEEEQKQRTSLASLKKKLEIENVD 377 + A KAQ +R+L ++DE ++E+ ++L K++R+ EA LE+E+KQR + KKK+EI+ D Sbjct: 1569 MQAMKAQFERDLQTRDEQNEEKKRLLLKQVRELEAELEDERKQRALAVASKKKMEIDLKD 1628 Query: 378 LMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRLRAAESS 557 L Q +Q++K Q + QRE++E +++E Q KE EK+L++ E+ Sbjct: 1629 LEAQIEAANKARDEVIKQLRKLQAQMKDYQRELEEARASRDEIFAQSKESEKKLKSLEAE 1688 Query: 558 TNQLIEDLSQSERIKRTLISERDDALEEIATLTVNKDTAIADKKRIESNLSALNMDYXXX 737 QL E+L+ SER +R ERD+ +EIA K + +K+R+E+ ++ L + Sbjct: 1689 ILQLQEELASSERARRHAEQERDELADEIANSASGKSALLDEKRRLEARIAQLEEELEEE 1748 Query: 738 XXXXXXXXDKNKKVTNQLEQVQNELNVERQNATRLENQKSTAERLNXXXXXXXXXXXXXT 917 D+ +K T Q++ + EL ER A + +N + ER N Sbjct: 1749 QSNMELLNDRFRKTTLQVDTLNTELAAERSAAQKSDNARQQLERQNKELKAKLQELEGAV 1808 Query: 918 GKKYKATVSAQQIKIQSLDAQLDAKIEEINQAXXXXXXXXXXXXXIMALMVDEQRRADNA 1097 K+KAT+SA + KI L+ QL+ + +E A I + DE+R AD Sbjct: 1809 KSKFKATISALEAKIGQLEEQLEQEAKERAAANKLVRRTEKKLKEIFMQVEDERRHADQY 1868 Query: 1098 IEQIEKATGRYKKAERSLQMEQENNSALTTQNRRLQRDLEEVNETKEARDQEIKVLKTKV 1277 EQ+EKA R K+ +R L+ +E + R+LQR+L++ E E +E+ LK ++ Sbjct: 1869 KEQMEKANARMKQLKRQLEEAEEEATRANASRRKLQRELDDATEANEGLSREVSTLKNRL 1928 Query: 1278 ER 1283 R Sbjct: 1929 RR 1930
Score = 70.9 bits (172), Expect = 1e-11
Identities = 92/440 (20%), Positives = 177/440 (40%), Gaps = 5/440 (1%)
Frame = +3
Query: 9 KSLLNRQLN---ELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNK 179
K++L QL EL + +++ + + K +LE L + + +EE EE N LQ K
Sbjct: 889 KNILAEQLQAETELFAEAEEMRARLAAKKQ---ELEEILHDLESRVEEEEERNQILQNEK 945
Query: 180 ERIEMQLTATKAQLDRELSSKDEMHDEQFKILNK-KLRDTEATLEEEQKQRTSLASLKKK 356
++++ + + QLD E ++ ++ E+ K K + E L E+Q + KK
Sbjct: 946 KKMQAHIQDLEEQLDEEEGARQKLQLEKVTAEAKIKKMEEEVLLLEDQNSK--FIKEKKL 1003
Query: 357 LEIENVDLMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKR 536
+E + +Q ++ + +L R QEV+ I +LE+R
Sbjct: 1004 MEDRIAECSSQLAE------------EEEKAKNLAKIRNKQEVM---------ISDLEER 1042
Query: 537 LRAAESSTNQLIEDLSQSERIKRTLISERDDALEEIATLTVNKDTAIADKKRIESNLSAL 716
L+ E + +L E+ KR L E D ++IA L D + E L
Sbjct: 1043 LKKEEKTRQEL-------EKAKRKLDGETTDLQDQIAELQAQVDELKVQLTKKEEELQGA 1095
Query: 717 NMDYXXXXXXXXXXXDKNKKVTNQLEQVQNELNVERQNATRLENQK-STAERLNXXXXXX 893
+++ Q+ ++Q + E+ + + E QK +E L
Sbjct: 1096 LARGDDETLHKNNALKVARELQAQIAELQEDFESEKASRNKAEKQKRDLSEELEALKTEL 1155
Query: 894 XXXXXXXTGKKYKATVSAQQIKIQSLDAQLDAKIEEINQAXXXXXXXXXXXXXIMALMVD 1073
T +AQ Q L + + ++ E+ +A A + D
Sbjct: 1156 EDTLD---------TTAAQ----QELRTKREQEVAELKKALEDETKNHE------AQIQD 1196
Query: 1074 EQRRADNAIEQIEKATGRYKKAERSLQMEQENNSALTTQNRRLQRDLEEVNETKEARDQE 1253
++R A+E++ + + K+ + +L ++N L T N+ L +++ + + K + +
Sbjct: 1197 MRQRHATALEELSEQLEQAKRFKANL---EKNKQGLETDNKELACEVKVLQQVKAESEHK 1253
Query: 1254 IKVLKTKVERLERRVRTGGR 1313
K L +V+ L +V G R
Sbjct: 1254 RKKLDAQVQELHAKVSEGDR 1273
Score = 65.9 bits (159), Expect = 3e-10
Identities = 81/434 (18%), Positives = 171/434 (39%), Gaps = 32/434 (7%)
Frame = +3
Query: 99 QLENDLENCKKTIEELEEENATLQMNKERIEMQLTATKAQLDRELSSKDEMHDEQFKILN 278
QLE + + ++ EE EE L+ ++ QL TK ++D +L + + + + + K+L
Sbjct: 1340 QLEEEKNSLQEQQEEEEEARKNLEKQVLALQSQLADTKKKVDDDLGTIESLEEAKKKLLK 1399
Query: 279 K----------------KLRDTEATLEEE--------QKQRTSLASLKKKLEIENVDLMN 386
KL T+ L++E QR +++L+KK + + L
Sbjct: 1400 DVEALSQRLEEKVLAYDKLEKTKNRLQQELDDLTVDLDHQRQIVSNLEKKQKKFDQLLAE 1459
Query: 387 QXXXXXXXXXXXXR---QIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRLRAAESS 557
+ R + ++ + +L+ R ++E L AK E +Q K+L + SS
Sbjct: 1460 EKGISARYAEERDRAEAEAREKETKALSLARALEEALEAKEEFERQNKQLRADMEDLMSS 1519
Query: 558 TNQLIEDLSQSERIKRTLISERDDALEEIATLTVNKDTAIADKKRIESNLSALNMDYXXX 737
+ + +++ + E+ KR L + ++ ++ L K R+E N+ A+ +
Sbjct: 1520 KDDVGKNVHELEKSKRALEQQVEEMRTQLEELEDELQATEDAKLRLEVNMQAMKAQFERD 1579
Query: 738 XXXXXXXXDKNKKVT-NQLEQVQNELNVERQNATRLENQKSTAERLNXXXXXXXXXXXXX 914
++ K++ Q+ +++ EL ER+ + + A +
Sbjct: 1580 LQTRDEQNEEKKRLLLKQVRELEAELEDERK-----QRALAVASKKKMEIDLKDLEAQIE 1634
Query: 915 TGKKYKATVSAQQIKIQSLDAQLDAKIEEINQAXXXXXXXXXXXXXIMALMVDEQRRADN 1094
K + V Q K+Q+ ++EE + + + E +
Sbjct: 1635 AANKARDEVIKQLRKLQAQMKDYQRELEEARASRDEIFAQSKESEKKLKSLEAEILQLQE 1694
Query: 1095 AIEQIEKATGRYKKAERSLQMEQENN----SALTTQNRRLQRDLEEVNETKEARDQEIKV 1262
+ E+A ++ L E N+ SAL + RRL+ + ++ E E +++
Sbjct: 1695 ELASSERARRHAEQERDELADEIANSASGKSALLDEKRRLEARIAQLEEELEEEQSNMEL 1754
Query: 1263 LKTKVERLERRVRT 1304
L + + +V T
Sbjct: 1755 LNDRFRKTTLQVDT 1768
Score = 62.0 bits (149), Expect = 5e-09
Identities = 94/492 (19%), Positives = 201/492 (40%), Gaps = 54/492 (10%)
Frame = +3
Query: 9 KSLLNRQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKERI 188
K+ L + L ++ ++ + L+ K + E+ + ++EL + + + ++ R+
Sbjct: 1219 KANLEKNKQGLETDNKELACEVKVLQQVKAESEHKRKKLDAQVQELHAKVS--EGDRLRV 1276
Query: 189 EMQLTATKAQ--LDRELSSKDEMHDEQFKI------LNKKLRDTEATLEEEQKQRTSLAS 344
E+ A K Q LD + +E + K L +L+DT+ L+EE +Q+ +L+S
Sbjct: 1277 ELAEKANKLQNELDNVSTLLEEAEKKGIKFAKDAAGLESQLQDTQELLQEETRQKLNLSS 1336
Query: 345 LKKKLEIENVDLMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVL-------NAKNE 503
++LE E L Q +Q+ Q ++++++ + L AK +
Sbjct: 1337 RIRQLEEEKNSLQEQQEEEEEARKNLEKQVLALQSQLADTKKKVDDDLGTIESLEEAKKK 1396
Query: 504 ALKQIKELEKRLR-------AAESSTNQL---IEDLSQSERIKRTLIS-------ERDDA 632
LK ++ L +RL E + N+L ++DL+ +R ++S + D
Sbjct: 1397 LLKDVEALSQRLEEKVLAYDKLEKTKNRLQQELDDLTVDLDHQRQIVSNLEKKQKKFDQL 1456
Query: 633 LEE----IATLTVNKDTAIADKKRIESNLSALNMDYXXXXXXXXXXXDKNKKVTNQLEQV 800
L E A +D A A+ + E+ +L +NK++ +E +
Sbjct: 1457 LAEEKGISARYAEERDRAEAEAREKETKALSLARALEEALEAKEEFERQNKQLRADMEDL 1516
Query: 801 QNELNVERQNATRLENQKSTAERLNXXXXXXXXXXXXXTGKKYKATVSAQ---QIKIQSL 971
+ + +N LE K E+ + +AT A+ ++ +Q++
Sbjct: 1517 MSSKDDVGKNVHELEKSKRALEQ----QVEEMRTQLEELEDELQATEDAKLRLEVNMQAM 1572
Query: 972 DAQLDAKIE-EINQAXXXXXXXXXXXXXIMALMVDEQRRADNAIEQIEKATGRYKKAERS 1148
AQ + ++ Q + A + DE+++ A+ +K K E
Sbjct: 1573 KAQFERDLQTRDEQNEEKKRLLLKQVRELEAELEDERKQRALAVASKKKMEIDLKDLEAQ 1632
Query: 1149 LQMEQENNSALTTQNRRLQRDLE----EVNETKEARDQ----------EIKVLKTKVERL 1286
++ + + Q R+LQ ++ E+ E + +RD+ ++K L+ ++ +L
Sbjct: 1633 IEAANKARDEVIKQLRKLQAQMKDYQRELEEARASRDEIFAQSKESEKKLKSLEAEILQL 1692
Query: 1287 ERRVRTGGRANR 1322
+ + + RA R
Sbjct: 1693 QEELASSERARR 1704
>sp|Q9JLT0|MYH10_RAT Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B) Length = 1976 Score = 241 bits (614), Expect = 5e-63 Identities = 138/422 (32%), Positives = 227/422 (53%) Frame = +3 Query: 18 LNRQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKERIEMQ 197 L + +L+S+KDDVGKN+ L+ +K LE +E + +EELE+E + K R+E+ Sbjct: 1509 LRADMEDLMSSKDDVGKNVHELEKSKRALEQQVEEMRTQLEELEDELQATEDAKLRLEVN 1568 Query: 198 LTATKAQLDRELSSKDEMHDEQFKILNKKLRDTEATLEEEQKQRTSLASLKKKLEIENVD 377 + A KAQ +R+L ++DE ++E+ ++L K++R+ EA LE+E+KQR + KKK+EI+ D Sbjct: 1569 MQAMKAQFERDLQTRDEQNEEKKRLLLKQVRELEAELEDERKQRALAVASKKKMEIDLKD 1628 Query: 378 LMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRLRAAESS 557 L Q +Q++K Q + QRE++E +++E Q KE EK+L++ E+ Sbjct: 1629 LEAQIEAANKARDEVIKQLRKLQAQMKDYQRELEEARASRDEIFAQSKESEKKLKSLEAE 1688 Query: 558 TNQLIEDLSQSERIKRTLISERDDALEEIATLTVNKDTAIADKKRIESNLSALNMDYXXX 737 QL E+L+ SER +R ERD+ +EIA K + +K+R+E+ ++ L + Sbjct: 1689 ILQLQEELASSERARRHAEQERDELADEIANSASGKSALLDEKRRLEARIAQLEEELEEE 1748 Query: 738 XXXXXXXXDKNKKVTNQLEQVQNELNVERQNATRLENQKSTAERLNXXXXXXXXXXXXXT 917 D+ +K T Q++ + EL ER A + +N + ER N Sbjct: 1749 QSNMELLNDRFRKTTLQVDTLNTELAAERSAAQKSDNARQQLERQNKELKAKLQELEGAV 1808 Query: 918 GKKYKATVSAQQIKIQSLDAQLDAKIEEINQAXXXXXXXXXXXXXIMALMVDEQRRADNA 1097 K+KAT+SA + KI L+ QL+ + +E A I + DE+R AD Sbjct: 1809 KSKFKATISALEAKIGQLEEQLEQEAKERAAANKLVRRTEKKLKEIFMQVEDERRHADQY 1868 Query: 1098 IEQIEKATGRYKKAERSLQMEQENNSALTTQNRRLQRDLEEVNETKEARDQEIKVLKTKV 1277 EQ+EKA R K+ +R L+ +E + R+LQR+L++ E E +E+ LK ++ Sbjct: 1869 KEQMEKANARMKQLKRQLEEAEEEATRANASRRKLQRELDDATEANEGLSREVSTLKNRL 1928 Query: 1278 ER 1283 R Sbjct: 1929 RR 1930
Score = 70.5 bits (171), Expect = 1e-11
Identities = 89/460 (19%), Positives = 179/460 (38%), Gaps = 28/460 (6%)
Frame = +3
Query: 3 NSKSLLNRQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKE 182
N K + + +L D+ L+ K+ E ++ ++ + LE++N+ K+
Sbjct: 943 NEKKKMQAHIQDLEEQLDEEEGARQKLQLEKVTAEAKIKKMEEEVLLLEDQNSKFIKEKK 1002
Query: 183 RIEMQLTATKAQLDRELSSKDEMHDEQFKILNKK---LRDTEATLEEEQKQRTSLASLKK 353
+E ++ A+ +L+ ++E KI NK+ + D E L++E+K R L K+
Sbjct: 1003 LMEDRI----AECSSQLAEEEEKAKNLAKIRNKQEVMISDLEERLKKEEKTRQELEKAKR 1058
Query: 354 KLEIENVDLMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEK 533
KL+ E DL +Q Q+ K + + + KN ALK +EL+
Sbjct: 1059 KLDGETTDLQDQIAELQAQVDELKVQLTKKEEELQGALARGDDETLHKNNALKVARELQA 1118
Query: 534 RL----------RAAESSTNQLIEDLSQSERIKRTLISERDDALEEIATLTVNKDTAIAD 683
++ +A+ + + DLS+ +T + + D L ++ +A+
Sbjct: 1119 QIAELQEDFESEKASRNKAEKQKRDLSEELEALKTELEDTLDTTAAQQELRTKREQEVAE 1178
Query: 684 -KKRIESNLSALNMDYXXXXXXXXXXXDKNKKVTNQLEQVQNELNVERQNATRLENQKST 860
KK +E D ++ LE++ +L ++ LE K
Sbjct: 1179 LKKALEDETK----------NHEAQIQDMRQRHATALEELSEQLEQAKRFKANLEKNKQG 1228
Query: 861 AERLNXXXXXXXXXXXXXTGKKYKATVSA-QQIKIQS--LDAQLDAKIEEINQAXXXXXX 1031
E N K+ V QQ+K +S +LDA+++E++
Sbjct: 1229 LETDN---------------KELACEVKVLQQVKAESEHKRKKLDAQVQELHAKVSEGDR 1273
Query: 1032 XXXXXXXIMALMVDEQRRADNAIEQIEKATGRYKK-----------AERSLQMEQENNSA 1178
+ +E +E+ EK ++ K + LQ E
Sbjct: 1274 LRVELAEKANKLQNELDNVSTLLEEAEKKGMKFAKDAAGLESQLQDTQELLQEETRQKLN 1333
Query: 1179 LTTQNRRLQRDLEEVNETKEARDQEIKVLKTKVERLERRV 1298
L+++ R+L+ + + E +E ++ K L+ +V L+ ++
Sbjct: 1334 LSSRIRQLEEEKNSLQEQQEEEEEARKNLEKQVLALQSQL 1373
Score = 65.1 bits (157), Expect = 5e-10
Identities = 81/434 (18%), Positives = 171/434 (39%), Gaps = 32/434 (7%)
Frame = +3
Query: 99 QLENDLENCKKTIEELEEENATLQMNKERIEMQLTATKAQLDRELSSKDEMHDEQFKILN 278
QLE + + ++ EE EE L+ ++ QL TK ++D +L + + + + + K+L
Sbjct: 1340 QLEEEKNSLQEQQEEEEEARKNLEKQVLALQSQLADTKKKVDDDLGTIEGLEEAKKKLLK 1399
Query: 279 K----------------KLRDTEATLEEE--------QKQRTSLASLKKKLEIENVDLMN 386
KL T+ L++E QR +++L+KK + + L
Sbjct: 1400 DVEALSQRLEEKVLAYDKLEKTKNRLQQELDDLTVDLDHQRQIVSNLEKKQKKFDQLLAE 1459
Query: 387 QXXXXXXXXXXXXR---QIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRLRAAESS 557
+ R + ++ + +L+ R ++E L AK E +Q K+L + SS
Sbjct: 1460 EKGISARYAEERDRAEAEAREKETKALSLARALEEALEAKEEFERQNKQLRADMEDLMSS 1519
Query: 558 TNQLIEDLSQSERIKRTLISERDDALEEIATLTVNKDTAIADKKRIESNLSALNMDYXXX 737
+ + +++ + E+ KR L + ++ ++ L K R+E N+ A+ +
Sbjct: 1520 KDDVGKNVHELEKSKRALEQQVEEMRTQLEELEDELQATEDAKLRLEVNMQAMKAQFERD 1579
Query: 738 XXXXXXXXDKNKKVT-NQLEQVQNELNVERQNATRLENQKSTAERLNXXXXXXXXXXXXX 914
++ K++ Q+ +++ EL ER+ + + A +
Sbjct: 1580 LQTRDEQNEEKKRLLLKQVRELEAELEDERK-----QRALAVASKKKMEIDLKDLEAQIE 1634
Query: 915 TGKKYKATVSAQQIKIQSLDAQLDAKIEEINQAXXXXXXXXXXXXXIMALMVDEQRRADN 1094
K + V Q K+Q+ ++EE + + + E +
Sbjct: 1635 AANKARDEVIKQLRKLQAQMKDYQRELEEARASRDEIFAQSKESEKKLKSLEAEILQLQE 1694
Query: 1095 AIEQIEKATGRYKKAERSLQMEQENN----SALTTQNRRLQRDLEEVNETKEARDQEIKV 1262
+ E+A ++ L E N+ SAL + RRL+ + ++ E E +++
Sbjct: 1695 ELASSERARRHAEQERDELADEIANSASGKSALLDEKRRLEARIAQLEEELEEEQSNMEL 1754
Query: 1263 LKTKVERLERRVRT 1304
L + + +V T
Sbjct: 1755 LNDRFRKTTLQVDT 1768
Score = 62.0 bits (149), Expect = 5e-09
Identities = 94/492 (19%), Positives = 201/492 (40%), Gaps = 54/492 (10%)
Frame = +3
Query: 9 KSLLNRQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKERI 188
K+ L + L ++ ++ + L+ K + E+ + ++EL + + + ++ R+
Sbjct: 1219 KANLEKNKQGLETDNKELACEVKVLQQVKAESEHKRKKLDAQVQELHAKVS--EGDRLRV 1276
Query: 189 EMQLTATKAQ--LDRELSSKDEMHDEQFKI------LNKKLRDTEATLEEEQKQRTSLAS 344
E+ A K Q LD + +E + K L +L+DT+ L+EE +Q+ +L+S
Sbjct: 1277 ELAEKANKLQNELDNVSTLLEEAEKKGMKFAKDAAGLESQLQDTQELLQEETRQKLNLSS 1336
Query: 345 LKKKLEIENVDLMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVL-------NAKNE 503
++LE E L Q +Q+ Q ++++++ + L AK +
Sbjct: 1337 RIRQLEEEKNSLQEQQEEEEEARKNLEKQVLALQSQLADTKKKVDDDLGTIEGLEEAKKK 1396
Query: 504 ALKQIKELEKRLR-------AAESSTNQL---IEDLSQSERIKRTLIS-------ERDDA 632
LK ++ L +RL E + N+L ++DL+ +R ++S + D
Sbjct: 1397 LLKDVEALSQRLEEKVLAYDKLEKTKNRLQQELDDLTVDLDHQRQIVSNLEKKQKKFDQL 1456
Query: 633 LEE----IATLTVNKDTAIADKKRIESNLSALNMDYXXXXXXXXXXXDKNKKVTNQLEQV 800
L E A +D A A+ + E+ +L +NK++ +E +
Sbjct: 1457 LAEEKGISARYAEERDRAEAEAREKETKALSLARALEEALEAKEEFERQNKQLRADMEDL 1516
Query: 801 QNELNVERQNATRLENQKSTAERLNXXXXXXXXXXXXXTGKKYKATVSAQ---QIKIQSL 971
+ + +N LE K E+ + +AT A+ ++ +Q++
Sbjct: 1517 MSSKDDVGKNVHELEKSKRALEQ----QVEEMRTQLEELEDELQATEDAKLRLEVNMQAM 1572
Query: 972 DAQLDAKIE-EINQAXXXXXXXXXXXXXIMALMVDEQRRADNAIEQIEKATGRYKKAERS 1148
AQ + ++ Q + A + DE+++ A+ +K K E
Sbjct: 1573 KAQFERDLQTRDEQNEEKKRLLLKQVRELEAELEDERKQRALAVASKKKMEIDLKDLEAQ 1632
Query: 1149 LQMEQENNSALTTQNRRLQRDLE----EVNETKEARDQ----------EIKVLKTKVERL 1286
++ + + Q R+LQ ++ E+ E + +RD+ ++K L+ ++ +L
Sbjct: 1633 IEAANKARDEVIKQLRKLQAQMKDYQRELEEARASRDEIFAQSKESEKKLKSLEAEILQL 1692
Query: 1287 ERRVRTGGRANR 1322
+ + + RA R
Sbjct: 1693 QEELASSERARR 1704
>sp|Q27991|MYH10_BOVIN Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B) Length = 1976 Score = 241 bits (614), Expect = 5e-63 Identities = 138/422 (32%), Positives = 227/422 (53%) Frame = +3 Query: 18 LNRQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKERIEMQ 197 L + +L+S+KDDVGKN+ L+ +K LE +E + +EELE+E + K R+E+ Sbjct: 1509 LRADMEDLMSSKDDVGKNVHELEKSKRALEQQVEEMRTQLEELEDELQATEDAKLRLEVN 1568 Query: 198 LTATKAQLDRELSSKDEMHDEQFKILNKKLRDTEATLEEEQKQRTSLASLKKKLEIENVD 377 + A KAQ +R+L ++DE ++E+ ++L K++R+ EA LE+E+KQR + KKK+EI+ D Sbjct: 1569 MQAMKAQFERDLQTRDEQNEEKKRLLIKQVRELEAELEDERKQRALAVASKKKMEIDLKD 1628 Query: 378 LMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRLRAAESS 557 L Q +Q++K Q + QRE++E +++E Q KE EK+L++ E+ Sbjct: 1629 LEAQIEAANKARDEVIKQLRKLQAQMKDYQRELEEARASRDEIFAQSKESEKKLKSLEAE 1688 Query: 558 TNQLIEDLSQSERIKRTLISERDDALEEIATLTVNKDTAIADKKRIESNLSALNMDYXXX 737 QL E+L+ SER +R ERD+ +EIA K + +K+R+E+ ++ L + Sbjct: 1689 ILQLQEELASSERARRHAEQERDELADEIANSASGKSALLDEKRRLEARIAQLEEELEEE 1748 Query: 738 XXXXXXXXDKNKKVTNQLEQVQNELNVERQNATRLENQKSTAERLNXXXXXXXXXXXXXT 917 D+ +K T Q++ + EL ER A + +N + ER N Sbjct: 1749 QSNMELLNDRFRKTTLQVDTLNTELAAERSAAQKSDNARQQLERQNKELKAKLQELEGAV 1808 Query: 918 GKKYKATVSAQQIKIQSLDAQLDAKIEEINQAXXXXXXXXXXXXXIMALMVDEQRRADNA 1097 K+KAT+SA + KI L+ QL+ + +E A I + DE+R AD Sbjct: 1809 KSKFKATISALEAKIGQLEEQLEQEAKERAAANKLVRRTEKKLKEIFMQVEDERRHADQY 1868 Query: 1098 IEQIEKATGRYKKAERSLQMEQENNSALTTQNRRLQRDLEEVNETKEARDQEIKVLKTKV 1277 EQ+EKA R K+ +R L+ +E + R+LQR+L++ E E +E+ LK ++ Sbjct: 1869 KEQMEKANARMKQLKRQLEEAEEEATRANASRRKLQRELDDATEANEGLSREVSTLKNRL 1928 Query: 1278 ER 1283 R Sbjct: 1929 RR 1930
Score = 72.0 bits (175), Expect = 4e-12
Identities = 82/434 (18%), Positives = 175/434 (40%), Gaps = 32/434 (7%)
Frame = +3
Query: 99 QLENDLENCKKTIEELEEENATLQMNKERIEMQLTATKAQLDRELSSKDEMHDEQFKILN 278
QLE + + ++ EE EE +L+ + ++ QLT TK ++D +L + + + + + K+L
Sbjct: 1340 QLEEERSSLQEQQEEEEEARRSLEKQLQALQAQLTDTKKKVDDDLGTIENLEEAKKKLLK 1399
Query: 279 K----------------KLRDTEATLEEE--------QKQRTSLASLKKKLEIENVDLMN 386
KL T+ L++E QR +++L+KK + + L
Sbjct: 1400 DVEVLSQRLEEKALAYDKLEKTKTRLQQELDDLLVDLDHQRQIVSNLEKKQKKFDQLLAE 1459
Query: 387 QXXXXXXXXXXXXR---QIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRLRAAESS 557
+ R + ++ + +L+ R ++E L A+ EA +Q K+L + SS
Sbjct: 1460 EKNISARYAEERDRAEAEAREKETKALSLARALEEALEAREEAERQNKQLRADMEDLMSS 1519
Query: 558 TNQLIEDLSQSERIKRTLISERDDALEEIATLTVNKDTAIADKKRIESNLSALNMDYXXX 737
+ + +++ + E+ KR L + ++ ++ L K R+E N+ A+ +
Sbjct: 1520 KDDVGKNVHELEKSKRALEQQVEEMRTQLEELEDELQATEDAKLRLEVNMQAMKAQFERD 1579
Query: 738 XXXXXXXXDKNKKVT-NQLEQVQNELNVERQNATRLENQKSTAERLNXXXXXXXXXXXXX 914
++ K++ Q+ +++ EL ER+ + + A +
Sbjct: 1580 LQTRDEQNEEKKRLLIKQVRELEAELEDERK-----QRALAVASKKKMEIDLKDLEAQIE 1634
Query: 915 TGKKYKATVSAQQIKIQSLDAQLDAKIEEINQAXXXXXXXXXXXXXIMALMVDEQRRADN 1094
K + V Q K+Q+ ++EE + + + E +
Sbjct: 1635 AANKARDEVIKQLRKLQAQMKDYQRELEEARASRDEIFAQSKESEKKLKSLEAEILQLQE 1694
Query: 1095 AIEQIEKATGRYKKAERSLQMEQENN----SALTTQNRRLQRDLEEVNETKEARDQEIKV 1262
+ E+A ++ L E N+ SAL + RRL+ + ++ E E +++
Sbjct: 1695 ELASSERARRHAEQERDELADEIANSASGKSALLDEKRRLEARIAQLEEELEEEQSNMEL 1754
Query: 1263 LKTKVERLERRVRT 1304
L + + +V T
Sbjct: 1755 LNDRFRKTTLQVDT 1768
Score = 71.6 bits (174), Expect = 6e-12
Identities = 88/439 (20%), Positives = 176/439 (40%), Gaps = 4/439 (0%)
Frame = +3
Query: 9 KSLLNRQLN---ELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNK 179
K++L QL EL + +++ + + K +LE L + + +EE EE N LQ K
Sbjct: 889 KNILAEQLQAETELFAEAEEMRARLAAKKQ---ELEEILHDLESRVEEEEERNQILQNEK 945
Query: 180 ERIEMQLTATKAQLDRELSSKDEMHDEQFKILNKKLRDTEATLEEEQKQRTSLASLKKKL 359
++++ + + QLD E ++ ++ E+ K++ E + + Q + KK +
Sbjct: 946 KKMQAHIQDLEEQLDEEEGARQKLQLEKV-TAEAKIKKMEEEILLLEDQNSKFIKEKKLM 1004
Query: 360 EIENVDLMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRL 539
E + +Q ++ + +L R QEV+ I +LE+RL
Sbjct: 1005 EDRIAECSSQLAE------------EEEKAKNLAKIRNKQEVM---------ISDLEERL 1043
Query: 540 RAAESSTNQLIEDLSQSERIKRTLISERDDALEEIATLTVNKDTAIADKKRIESNLSALN 719
+ E + +L E+ KR L E D ++IA L D + E L
Sbjct: 1044 KKEEKTRQEL-------EKAKRKLDGETTDLQDQIAELQAQIDELKIQVAKKEEELQGAL 1096
Query: 720 MDYXXXXXXXXXXXDKNKKVTNQLEQVQNELNVERQNATRLENQK-STAERLNXXXXXXX 896
+++ Q+ ++Q + E+ + + E QK +E L
Sbjct: 1097 ARGDDETLHKNNALKVVRELQAQIAELQEDFESEKASRNKAEKQKRDLSEELEALKTELE 1156
Query: 897 XXXXXXTGKKYKATVSAQQIKIQSLDAQLDAKIEEINQAXXXXXXXXXXXXXIMALMVDE 1076
++ T Q++ A+L +EE ++ A + D
Sbjct: 1157 DTLDTTAAQQELRTKREQEV------AELKKALEEETKSHE-------------AQIQDM 1197
Query: 1077 QRRADNAIEQIEKATGRYKKAERSLQMEQENNSALTTQNRRLQRDLEEVNETKEARDQEI 1256
++R A+E++ + + K+ + +L ++N L T N+ L +++ + + K + +
Sbjct: 1198 RQRHATALEELSEQLEQAKRFKANL---EKNKQGLETDNKELACEVKVLQQVKAESEHKR 1254
Query: 1257 KVLKTKVERLERRVRTGGR 1313
K L +V+ L +V G R
Sbjct: 1255 KKLDAQVQELHAKVSEGDR 1273
Score = 62.8 bits (151), Expect = 3e-09
Identities = 91/492 (18%), Positives = 198/492 (40%), Gaps = 54/492 (10%)
Frame = +3
Query: 9 KSLLNRQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKERI 188
K+ L + L ++ ++ + L+ K + E+ + ++EL + + + ++ R+
Sbjct: 1219 KANLEKNKQGLETDNKELACEVKVLQQVKAESEHKRKKLDAQVQELHAKVS--EGDRLRV 1276
Query: 189 EMQLTATKAQ--LDRELSSKDEMHDEQFKI------LNKKLRDTEATLEEEQKQRTSLAS 344
E+ A K Q LD + +E + K L +L+DT+ L+EE +Q+ +L+S
Sbjct: 1277 ELAEKANKLQNELDNVSTLLEEAEKKGIKFAKDAAGLESQLQDTQELLQEETRQKLNLSS 1336
Query: 345 LKKKLEIENVDLMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVL-------NAKNE 503
++LE E L Q +Q++ Q ++++++ + L AK +
Sbjct: 1337 RIRQLEEERSSLQEQQEEEEEARRSLEKQLQALQAQLTDTKKKVDDDLGTIENLEEAKKK 1396
Query: 504 ALKQIKELEKRLR--------------AAESSTNQLIEDLSQSERIKRTLISER---DDA 632
LK ++ L +RL + + L+ DL +I L ++ D
Sbjct: 1397 LLKDVEVLSQRLEEKALAYDKLEKTKTRLQQELDDLLVDLDHQRQIVSNLEKKQKKFDQL 1456
Query: 633 LEEIATLTV----NKDTAIADKKRIESNLSALNMDYXXXXXXXXXXXDKNKKVTNQLEQV 800
L E ++ +D A A+ + E+ +L +NK++ +E +
Sbjct: 1457 LAEEKNISARYAEERDRAEAEAREKETKALSLARALEEALEAREEAERQNKQLRADMEDL 1516
Query: 801 QNELNVERQNATRLENQKSTAERLNXXXXXXXXXXXXXTGKKYKATVSAQ---QIKIQSL 971
+ + +N LE K E+ + +AT A+ ++ +Q++
Sbjct: 1517 MSSKDDVGKNVHELEKSKRALEQ----QVEEMRTQLEELEDELQATEDAKLRLEVNMQAM 1572
Query: 972 DAQLDAKIE-EINQAXXXXXXXXXXXXXIMALMVDEQRRADNAIEQIEKATGRYKKAERS 1148
AQ + ++ Q + A + DE+++ A+ +K K E
Sbjct: 1573 KAQFERDLQTRDEQNEEKKRLLIKQVRELEAELEDERKQRALAVASKKKMEIDLKDLEAQ 1632
Query: 1149 LQMEQENNSALTTQNRRLQRDLE----EVNETKEARDQ----------EIKVLKTKVERL 1286
++ + + Q R+LQ ++ E+ E + +RD+ ++K L+ ++ +L
Sbjct: 1633 IEAANKARDEVIKQLRKLQAQMKDYQRELEEARASRDEIFAQSKESEKKLKSLEAEILQL 1692
Query: 1287 ERRVRTGGRANR 1322
+ + + RA R
Sbjct: 1693 QEELASSERARR 1704
>sp|P35580|MYH10_HUMAN Myosin-10 (Myosin heavy chain, nonmuscle IIb) (Nonmuscle myosin heavy chain IIb) (NMMHC II-b) (NMMHC-IIB) (Cellular myosin heavy chain, type B) (Nonmuscle myosin heavy chain-B) (NMMHC-B) Length = 1976 Score = 238 bits (608), Expect = 3e-62 Identities = 137/422 (32%), Positives = 226/422 (53%) Frame = +3 Query: 18 LNRQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKERIEMQ 197 L + +L+S+KDDVGKN+ L+ +K LE +E + +EELE+E + K R+E+ Sbjct: 1509 LRADMEDLMSSKDDVGKNVHELEKSKRALEQQVEEMRTQLEELEDELQATEDAKLRLEVN 1568 Query: 198 LTATKAQLDRELSSKDEMHDEQFKILNKKLRDTEATLEEEQKQRTSLASLKKKLEIENVD 377 + A KAQ +R+L ++DE ++E+ ++L K++R+ EA LE+E+KQR + KKK+EI+ D Sbjct: 1569 MQAMKAQFERDLQTRDEQNEEKKRLLIKQVRELEAELEDERKQRALAVASKKKMEIDLKD 1628 Query: 378 LMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRLRAAESS 557 L Q +Q++K Q + QRE++E +++E Q KE EK+L++ E+ Sbjct: 1629 LEAQIEAANKARDEVIKQLRKLQAQMKDYQRELEEARASRDEIFAQSKESEKKLKSLEAE 1688 Query: 558 TNQLIEDLSQSERIKRTLISERDDALEEIATLTVNKDTAIADKKRIESNLSALNMDYXXX 737 QL E+L+ SER +R ERD+ +EI K + +K+R+E+ ++ L + Sbjct: 1689 ILQLQEELASSERARRHAEQERDELADEITNSASGKSALLDEKRRLEARIAQLEEELEEE 1748 Query: 738 XXXXXXXXDKNKKVTNQLEQVQNELNVERQNATRLENQKSTAERLNXXXXXXXXXXXXXT 917 D+ +K T Q++ + EL ER A + +N + ER N Sbjct: 1749 QSNMELLNDRFRKTTLQVDTLNAELAAERSAAQKSDNARQQLERQNKELKAKLQELEGAV 1808 Query: 918 GKKYKATVSAQQIKIQSLDAQLDAKIEEINQAXXXXXXXXXXXXXIMALMVDEQRRADNA 1097 K+KAT+SA + KI L+ QL+ + +E A I + DE+R AD Sbjct: 1809 KSKFKATISALEAKIGQLEEQLEQEAKERAAANKLVRRTEKKLKEIFMQVEDERRHADQY 1868 Query: 1098 IEQIEKATGRYKKAERSLQMEQENNSALTTQNRRLQRDLEEVNETKEARDQEIKVLKTKV 1277 EQ+EKA R K+ +R L+ +E + R+LQR+L++ E E +E+ LK ++ Sbjct: 1869 KEQMEKANARMKQLKRQLEEAEEEATRANASRRKLQRELDDATEANEGLSREVSTLKNRL 1928 Query: 1278 ER 1283 R Sbjct: 1929 RR 1930
Score = 70.5 bits (171), Expect = 1e-11
Identities = 88/439 (20%), Positives = 175/439 (39%), Gaps = 4/439 (0%)
Frame = +3
Query: 9 KSLLNRQLN---ELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNK 179
K++L QL EL + +++ + + K +LE L + + +EE EE N LQ K
Sbjct: 889 KNILAEQLQAETELFAEAEEMRARLAAKKQ---ELEEILHDLESRVEEEEERNQILQNEK 945
Query: 180 ERIEMQLTATKAQLDRELSSKDEMHDEQFKILNKKLRDTEATLEEEQKQRTSLASLKKKL 359
++++ + + QLD E ++ ++ E+ K++ E + + Q + KK +
Sbjct: 946 KKMQAHIQDLEEQLDEEEGARQKLQLEKV-TAEAKIKKMEEEILLLEDQNSKFIKEKKLM 1004
Query: 360 EIENVDLMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRL 539
E + +Q ++ + +L R QEV+ I +LE+RL
Sbjct: 1005 EDRIAECSSQLAE------------EEEKAKNLAKIRNKQEVM---------ISDLEERL 1043
Query: 540 RAAESSTNQLIEDLSQSERIKRTLISERDDALEEIATLTVNKDTAIADKKRIESNLSALN 719
+ E + +L E+ KR L E D ++IA L D + E L
Sbjct: 1044 KKEEKTRQEL-------EKAKRKLDGETTDLQDQIAELQAQIDELKLQLAKKEEELQGAL 1096
Query: 720 MDYXXXXXXXXXXXDKNKKVTNQLEQVQNELNVERQNATRLENQK-STAERLNXXXXXXX 896
+++ Q+ ++Q + E+ + + E QK +E L
Sbjct: 1097 ARGDDETLHKNNALKVVRELQAQIAELQEDFESEKASRNKAEKQKRDLSEELEALKTELE 1156
Query: 897 XXXXXXTGKKYKATVSAQQIKIQSLDAQLDAKIEEINQAXXXXXXXXXXXXXIMALMVDE 1076
++ T Q++ A+L +EE + A + D
Sbjct: 1157 DTLDTTAAQQELRTKREQEV------AELKKALEEETKNHE-------------AQIQDM 1197
Query: 1077 QRRADNAIEQIEKATGRYKKAERSLQMEQENNSALTTQNRRLQRDLEEVNETKEARDQEI 1256
++R A+E++ + + K+ + +L ++N L T N+ L +++ + + K + +
Sbjct: 1198 RQRHATALEELSEQLEQAKRFKANL---EKNKQGLETDNKELACEVKVLQQVKAESEHKR 1254
Query: 1257 KVLKTKVERLERRVRTGGR 1313
K L +V+ L +V G R
Sbjct: 1255 KKLDAQVQELHAKVSEGDR 1273
Score = 66.6 bits (161), Expect = 2e-10
Identities = 82/435 (18%), Positives = 167/435 (38%), Gaps = 6/435 (1%)
Frame = +3
Query: 18 LNRQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKERIEMQ 197
L QL + DD I SL+ AK +L D E + +EE L+ K R++ +
Sbjct: 1369 LQSQLADTKKKVDDDLGTIESLEEAKKKLLKDAEALSQRLEEKALAYDKLEKTKNRLQQE 1428
Query: 198 LTATKAQLDRELSSKDEMHDEQFKILNKKLRDTEATLEEEQKQRTSLASLKKKLEIENVD 377
L LD + R + LE++QK+ L + +K + +
Sbjct: 1429 LDDLTVDLDHQ-------------------RQVASNLEKKQKKFDQLLAEEKSISARYAE 1469
Query: 378 LMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRLRAAESS 557
++ + ++ + +L+ R ++E L AK E +Q K+L + SS
Sbjct: 1470 ERDRAEA----------EAREKETKALSLARALEEALEAKEEFERQNKQLRADMEDLMSS 1519
Query: 558 TNQLIEDLSQSERIKRTLISERDDALEEIATLTVNKDTAIADKKRIESNLSALNMDYXXX 737
+ + +++ + E+ KR L + ++ ++ L K R+E N+ A+ +
Sbjct: 1520 KDDVGKNVHELEKSKRALEQQVEEMRTQLEELEDELQATEDAKLRLEVNMQAMKAQFERD 1579
Query: 738 XXXXXXXXDKNKKVT-NQLEQVQNELNVERQNATRLENQKSTAERLNXXXXXXXXXXXXX 914
++ K++ Q+ +++ EL ER+ + + A +
Sbjct: 1580 LQTRDEQNEEKKRLLIKQVRELEAELEDERK-----QRALAVASKKKMEIDLKDLEAQIE 1634
Query: 915 TGKKYKATVSAQQIKIQSLDAQLDAKIEEINQAXXXXXXXXXXXXXIMALMVDEQRRADN 1094
K + V Q K+Q+ ++EE + + + E +
Sbjct: 1635 AANKARDEVIKQLRKLQAQMKDYQRELEEARASRDEIFAQSKESEKKLKSLEAEILQLQE 1694
Query: 1095 AIEQIEKATGRYKKAERSLQMEQENNSA-----LTTQNRRLQRDLEEVNETKEARDQEIK 1259
+ E+A R+ + ER ++ NSA L + RRL+ + ++ E E ++
Sbjct: 1695 ELASSERAR-RHAEQERDELADEITNSASGKSALLDEKRRLEARIAQLEEELEEEQSNME 1753
Query: 1260 VLKTKVERLERRVRT 1304
+L + + +V T
Sbjct: 1754 LLNDRFRKTTLQVDT 1768
Score = 60.8 bits (146), Expect = 1e-08
Identities = 90/494 (18%), Positives = 198/494 (40%), Gaps = 56/494 (11%)
Frame = +3
Query: 9 KSLLNRQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKERI 188
K+ L + L ++ ++ + L+ K + E+ + ++EL + + +R+
Sbjct: 1219 KANLEKNKQGLETDNKELACEVKVLQQVKAESEHKRKKLDAQVQELHAKVS----EGDRL 1274
Query: 189 EMQLTATKAQLDRELSSKDEMHDEQFK----------ILNKKLRDTEATLEEEQKQRTSL 338
++L ++L EL + + +E K L +L+DT+ L+EE +Q+ +L
Sbjct: 1275 RVELAEKASKLQNELDNVSTLLEEAEKKGIKFAKDAASLESQLQDTQELLQEETRQKLNL 1334
Query: 339 ASLKKKLEIENVDLMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVL-------NAK 497
+S ++LE E L Q +Q+ Q ++++++ + L AK
Sbjct: 1335 SSRIRQLEEEKNSLQEQQEEEEEARKNLEKQVLALQSQLADTKKKVDDDLGTIESLEEAK 1394
Query: 498 NEALKQIKELEKRLR-------AAESSTNQLIE-------DLSQSERIKRTLISER---D 626
+ LK + L +RL E + N+L + DL ++ L ++ D
Sbjct: 1395 KKLLKDAEALSQRLEEKALAYDKLEKTKNRLQQELDDLTVDLDHQRQVASNLEKKQKKFD 1454
Query: 627 DALEEIATLTV----NKDTAIADKKRIESNLSALNMDYXXXXXXXXXXXDKNKKVTNQLE 794
L E +++ +D A A+ + E+ +L +NK++ +E
Sbjct: 1455 QLLAEEKSISARYAEERDRAEAEAREKETKALSLARALEEALEAKEEFERQNKQLRADME 1514
Query: 795 QVQNELNVERQNATRLENQKSTAERLNXXXXXXXXXXXXXTGKKYKATVSAQ---QIKIQ 965
+ + + +N LE K E+ + +AT A+ ++ +Q
Sbjct: 1515 DLMSSKDDVGKNVHELEKSKRALEQ----QVEEMRTQLEELEDELQATEDAKLRLEVNMQ 1570
Query: 966 SLDAQLDAKIE-EINQAXXXXXXXXXXXXXIMALMVDEQRRADNAIEQIEKATGRYKKAE 1142
++ AQ + ++ Q + A + DE+++ A+ +K K E
Sbjct: 1571 AMKAQFERDLQTRDEQNEEKKRLLIKQVRELEAELEDERKQRALAVASKKKMEIDLKDLE 1630
Query: 1143 RSLQMEQENNSALTTQNRRLQRDLE----EVNETKEARDQ----------EIKVLKTKVE 1280
++ + + Q R+LQ ++ E+ E + +RD+ ++K L+ ++
Sbjct: 1631 AQIEAANKARDEVIKQLRKLQAQMKDYQRELEEARASRDEIFAQSKESEKKLKSLEAEIL 1690
Query: 1281 RLERRVRTGGRANR 1322
+L+ + + RA R
Sbjct: 1691 QLQEELASSERARR 1704
>sp|P35579|MYH9_HUMAN Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A) Length = 1960 Score = 238 bits (607), Expect = 4e-62 Identities = 136/419 (32%), Positives = 229/419 (54%) Frame = +3 Query: 27 QLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKERIEMQLTA 206 ++ +L+S+KDDVGK++ L+ +K LE +E K +EELE+E + K R+E+ L A Sbjct: 1505 EMEDLMSSKDDVGKSVHELEKSKRALEQQVEEMKTQLEELEDELQATEDAKLRLEVNLQA 1564 Query: 207 TKAQLDRELSSKDEMHDEQFKILNKKLRDTEATLEEEQKQRTSLASLKKKLEIENVDLMN 386 KAQ +R+L +DE +E+ K L +++R+ EA LE+E+KQR+ + +KKLE++ DL Sbjct: 1565 MKAQFERDLQGRDEQSEEKKKQLVRQVREMEAELEDERKQRSMAVAARKKLEMDLKDLEA 1624 Query: 387 QXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRLRAAESSTNQ 566 +Q++K Q + RE+ + ++ E L Q KE EK+L++ E+ Q Sbjct: 1625 HIDSANKNRDEAIKQLRKLQAQMKDCMRELDDTRASREEILAQAKENEKKLKSMEAEMIQ 1684 Query: 567 LIEDLSQSERIKRTLISERDDALEEIATLTVNKDTAIADKKRIESNLSALNMDYXXXXXX 746 L E+L+ +ER KR ERD+ +EIA + A+ +K+R+E+ ++ L + Sbjct: 1685 LQEELAAAERAKRQAQQERDELADEIANSSGKGALALEEKRRLEARIAQLEEELEEEQGN 1744 Query: 747 XXXXXDKNKKVTNQLEQVQNELNVERQNATRLENQKSTAERLNXXXXXXXXXXXXXTGKK 926 D+ KK Q++Q+ +LN+ER +A + EN + ER N K Sbjct: 1745 TELINDRLKKANLQIDQINTDLNLERSHAQKNENARQQLERQNKELKVKLQEMEGTVKSK 1804 Query: 927 YKATVSAQQIKIQSLDAQLDAKIEEINQAXXXXXXXXXXXXXIMALMVDEQRRADNAIEQ 1106 YKA+++A + KI L+ QLD + +E A ++ + DE+R A+ +Q Sbjct: 1805 YKASITALEAKIAQLEEQLDNETKERQAACKQVRRTEKKLKDVLLQVDDERRNAEQYKDQ 1864 Query: 1107 IEKATGRYKKAERSLQMEQENNSALTTQNRRLQRDLEEVNETKEARDQEIKVLKTKVER 1283 +KA+ R K+ +R L+ +E R+LQR+LE+ ET +A ++E+ LK K+ R Sbjct: 1865 ADKASTRLKQLKRQLEEAEEEAQRANASRRKLQRELEDATETADAMNREVSSLKNKLRR 1923
Score = 75.1 bits (183), Expect = 5e-13
Identities = 87/452 (19%), Positives = 180/452 (39%), Gaps = 22/452 (4%)
Frame = +3
Query: 9 KSLLNRQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKERI 188
K + + + EL ++ L+ K+ E L+ ++ LE++N L K+ +
Sbjct: 938 KKKMQQNIQELEEQLEEEESARQKLQLEKVTTEAKLKKLEEEQIILEDQNCKLAKEKKLL 997
Query: 189 EMQLTATKAQLDRELSSKDEMHDEQFKILNKK---LRDTEATLEEEQKQRTSLASLKKKL 359
E ++ A+ L+ ++E K+ NK + D E L E+KQR L ++KL
Sbjct: 998 EDRI----AEFTTNLTEEEEKSKSLAKLKNKHEAMITDLEERLRREEKQRQELEKTRRKL 1053
Query: 360 EIENVDLMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRL 539
E ++ DL +Q Q+ K + + ++E KN ALK+I+ELE ++
Sbjct: 1054 EGDSTDLSDQIAELQAQIAELKMQLAKKEEELQAALARVEEEAAQKNMALKKIRELESQI 1113
Query: 540 RAAESSTNQLIEDLSQSERIKRTLISERDDALEEIATLTVNKDTA---IADKKRIESNLS 710
+ +++E+ KR L E +AL+ T++ A + K+ E N+
Sbjct: 1114 SELQEDLESERASRNKAEKQKRDL-GEELEALKTELEDTLDSTAAQQELRSKREQEVNIL 1172
Query: 711 ALNMDYXXXXXXXXXXXDKNKKVTNQLEQVQNELNVERQNATRLENQKSTAERLNXXXXX 890
++ + +K + +E++ +L ++ LE K T E
Sbjct: 1173 KKTLE-EEAKTHEAQIQEMRQKHSQAVEELAEQLEQTKRVKANLEKAKQTLENERGELAN 1231
Query: 891 XXXXXXXXTG------KKYKATVSAQQIKI---QSLDAQLDAKIEEINQAXXXXXXXXXX 1043
G KK +A + Q+K + + +L K+ ++
Sbjct: 1232 EVKVLLQGKGDSEHKRKKVEAQLQELQVKFNEGERVRTELADKVTKLQ----------VE 1281
Query: 1044 XXXIMALMVDEQRRADNAIEQIEKATGRYKKAERSLQMEQENNSALTTQNRRLQ------ 1205
+ L+ ++ + + + + LQ E +L+T+ ++++
Sbjct: 1282 LDNVTGLLSQSDSKSSKLTKDFSALESQLQDTQELLQEENRQKLSLSTKLKQVEDEKNSF 1341
Query: 1206 -RDLEEVNETKEARDQEIKVLKTKVERLERRV 1298
LEE E K +++I L +V +++++
Sbjct: 1342 REQLEEEEEAKHNLEKQIATLHAQVADMKKKM 1373
Score = 65.1 bits (157), Expect = 5e-10
Identities = 81/442 (18%), Positives = 177/442 (40%), Gaps = 21/442 (4%)
Frame = +3
Query: 6 SKSLLNRQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKER 185
+K L + EL + + + G ++ + ++E L+ + E E L +
Sbjct: 1218 AKQTLENERGELANEVKVLLQGKGDSEHKRKKVEAQLQELQVKFNEGERVRTELADKVTK 1277
Query: 186 IEMQLTATKAQLDRELSSKDEMHDEQFKILNKKLRDTEATLEEEQKQRTSLASLKKKLEI 365
++++L L + SK + F L +L+DT+ L+EE +Q+ SL++ K++E
Sbjct: 1278 LQVELDNVTGLLSQS-DSKSSKLTKDFSALESQLQDTQELLQEENRQKLSLSTKLKQVED 1336
Query: 366 ENVDLMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRLRA 545
E Q +QI + +++M++ + A + ++L+K L
Sbjct: 1337 EKNSFREQLEEEEEAKHNLEKQIATLHAQVADMKKKMEDSVGCLETAEEVKRKLQKDLEG 1396
Query: 546 AESSTNQLIEDLSQSERIKRTLISERDDALEEI-----ATLTVNK-----DTAIADKKRI 695
+ + + E+ K L E DD L ++ + + K D +A++K I
Sbjct: 1397 LSQRHEEKVAAYDKLEKTKTRLQQELDDLLVDLDHQRQSACNLEKKQKKFDQLLAEEKTI 1456
Query: 696 ESNLSALNMDYXXXXXXXXXXXDKNKKVTNQLEQVQNELNVERQNATRLENQKSTAERLN 875
+ + ++++ E+ L++ R +E QK+ ERLN
Sbjct: 1457 SAKYA----------------EERDRAEAEAREKETKALSLARALEEAME-QKAELERLN 1499
Query: 876 ---XXXXXXXXXXXXXTGK---KYKATVSAQQIKIQSLDAQLDAKIEEINQAXXXXXXXX 1037
GK + + + A + +++ + QL+ +E+
Sbjct: 1500 KQFRTEMEDLMSSKDDVGKSVHELEKSKRALEQQVEEMKTQLEELEDELQATEDAKLRLE 1559
Query: 1038 XXXXXIMALMV-DEQRRADNAIEQIEKATGRYKKAERSLQMEQENNSALTTQNRRLQRDL 1214
+ A D Q R + + E+ ++ + ++ E L+ E++ S ++L+ DL
Sbjct: 1560 VNLQAMKAQFERDLQGRDEQSEEKKKQLVRQVREMEAELEDERKQRSMAVAARKKLEMDL 1619
Query: 1215 EE----VNETKEARDQEIKVLK 1268
++ ++ + RD+ IK L+
Sbjct: 1620 KDLEAHIDSANKNRDEAIKQLR 1641
Score = 54.3 bits (129), Expect = 9e-07
Identities = 84/432 (19%), Positives = 153/432 (35%), Gaps = 49/432 (11%)
Frame = +3
Query: 138 EELEEENATLQMNKERIEMQLTATKAQLDRELSSKDEMHDEQFKI----------LNKKL 287
++L EN +M E ++ QL A K QL +L ++ E+ E ++ L +
Sbjct: 860 KQLAAENRLTEM--ETLQSQLMAEKLQLQEQLQAETELCAEAEELRARLTAKKQELEEIC 917
Query: 288 RDTEATLEEEQKQRTSLASLK-----------------------------------KKLE 362
D EA +EEE+++ L + K KKLE
Sbjct: 918 HDLEARVEEEEERCQHLQAEKKKMQQNIQELEEQLEEEESARQKLQLEKVTTEAKLKKLE 977
Query: 363 IENVDLMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRLR 542
E + L +Q +I + + + + + KN+ I +LE+RLR
Sbjct: 978 EEQIILEDQNCKLAKEKKLLEDRIAEFTTNLTEEEEKSKSLAKLKNKHEAMITDLEERLR 1037
Query: 543 AAESSTNQLIEDLSQSERIKRTLISERDDALEEIATLTVNKDTAIADKKRIESNLSALNM 722
E +L E+ +R L + D ++IA L + E L A
Sbjct: 1038 REEKQRQEL-------EKTRRKLEGDSTDLSDQIAELQAQIAELKMQLAKKEEELQAALA 1090
Query: 723 DYXXXXXXXXXXXDKNKKVTNQLEQVQNELNVERQNATRLENQK-STAERLNXXXXXXXX 899
K +++ +Q+ ++Q +L ER + + E QK E L
Sbjct: 1091 RVEEEAAQKNMALKKIRELESQISELQEDLESERASRNKAEKQKRDLGEELEALKTELED 1150
Query: 900 XXXXXTGKKYKATVSAQQIKIQSLDAQLDAKIEEINQAXXXXXXXXXXXXXIMALMVDEQ 1079
++ + Q++ I + +AK E I +
Sbjct: 1151 TLDSTAAQQELRSKREQEVNILKKTLEEEAKTHEAQ---------------IQEMRQKHS 1195
Query: 1080 RRADNAIEQIEKATGRYKKAERSLQMEQENNSALTTQNRRL---QRDLEEVNETKEARDQ 1250
+ + EQ+E+ E++ Q + L + + L + D E + EA+ Q
Sbjct: 1196 QAVEELAEQLEQTKRVKANLEKAKQTLENERGELANEVKVLLQGKGDSEHKRKKVEAQLQ 1255
Query: 1251 EIKVLKTKVERL 1286
E++V + ER+
Sbjct: 1256 ELQVKFNEGERV 1267
Score = 43.1 bits (100), Expect = 0.002
Identities = 49/221 (22%), Positives = 92/221 (41%), Gaps = 16/221 (7%)
Frame = +3
Query: 24 RQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKERIEMQLT 203
R + +D++ I + + + I +LEEE Q N E I +L
Sbjct: 1694 RAKRQAQQERDELADEIANSSGKGALALEEKRRLEARIAQLEEELEEEQGNTELINDRLK 1753
Query: 204 ATKAQLD---------RELSSKDEMHDEQFKILNK----KLRDTEATLEEEQKQRTSLAS 344
Q+D R + K+E +Q + NK KL++ E T+ + K + S+ +
Sbjct: 1754 KANLQIDQINTDLNLERSHAQKNENARQQLERQNKELKVKLQEMEGTV--KSKYKASITA 1811
Query: 345 LKKKLEIENVDLMNQXXXXXXXXXXXXRQIKKHQGTSL---NSQREMQEVLNAKNEALKQ 515
L+ K+ L N+ R KK + L + +R ++ + ++A +
Sbjct: 1812 LEAKIAQLEEQLDNETKERQAACKQVRRTEKKLKDVLLQVDDERRNAEQYKDQADKASTR 1871
Query: 516 IKELEKRLRAAESSTNQLIEDLSQSERIKRTLISERDDALE 638
+K+L+++L AE E+ ++ +R L E +DA E
Sbjct: 1872 LKQLKRQLEEAE-------EEAQRANASRRKLQRELEDATE 1905
Score = 33.5 bits (75), Expect = 1.7
Identities = 19/75 (25%), Positives = 39/75 (52%), Gaps = 3/75 (4%)
Frame = +3
Query: 24 RQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKERIEMQL- 200
++L +++ DD +N K+ + L+ K+ +EE EEE ++ +++ +L
Sbjct: 1842 KKLKDVLLQVDDERRNAEQYKDQADKASTRLKQLKRQLEEAEEEAQRANASRRKLQRELE 1901
Query: 201 --TATKAQLDRELSS 239
T T ++RE+SS
Sbjct: 1902 DATETADAMNREVSS 1916
>sp|Q8VDD5|MYH9_MOUSE Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A) Length = 1960 Score = 238 bits (607), Expect = 4e-62 Identities = 136/419 (32%), Positives = 229/419 (54%) Frame = +3 Query: 27 QLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKERIEMQLTA 206 ++ +L+S+KDDVGK++ L+ +K LE +E K +EELE+E + K R+E+ L A Sbjct: 1505 EMEDLMSSKDDVGKSVHELEKSKRALEQQVEEMKTQLEELEDELQATEDAKLRLEVNLQA 1564 Query: 207 TKAQLDRELSSKDEMHDEQFKILNKKLRDTEATLEEEQKQRTSLASLKKKLEIENVDLMN 386 KAQ +R+L +DE +E+ K L +++R+ EA LE+E+KQR+ + +KKLE++ DL Sbjct: 1565 MKAQFERDLQGRDEQSEEKKKQLVRQVREMEAELEDERKQRSMAMAARKKLEMDLKDLEA 1624 Query: 387 QXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRLRAAESSTNQ 566 +Q++K Q + RE+ + ++ E L Q KE EK+L++ E+ Q Sbjct: 1625 HIDTANKNREEAIKQLRKLQAQMKDCMRELDDTRASREEILAQAKENEKKLKSMEAEMIQ 1684 Query: 567 LIEDLSQSERIKRTLISERDDALEEIATLTVNKDTAIADKKRIESNLSALNMDYXXXXXX 746 L E+L+ +ER KR ERD+ +EIA + A+ +K+R+E+ ++ L + Sbjct: 1685 LQEELAAAERAKRQAQQERDELADEIANSSGKGALALEEKRRLEARIALLEEELEEEQGN 1744 Query: 747 XXXXXDKNKKVTNQLEQVQNELNVERQNATRLENQKSTAERLNXXXXXXXXXXXXXTGKK 926 D+ KK Q++Q+ +LN+ER +A + EN + ER N K Sbjct: 1745 TELINDRLKKANLQIDQINTDLNLERSHAQKNENARQQLERQNKELKAKLQEMESAVKSK 1804 Query: 927 YKATVSAQQIKIQSLDAQLDAKIEEINQAXXXXXXXXXXXXXIMALMVDEQRRADNAIEQ 1106 YKA+++A + KI L+ QLD + +E A ++ + DE+R A+ +Q Sbjct: 1805 YKASIAALEAKIAQLEEQLDNETKERQAASKQVRRTEKKLKDVLLQVEDERRNAEQFKDQ 1864 Query: 1107 IEKATGRYKKAERSLQMEQENNSALTTQNRRLQRDLEEVNETKEARDQEIKVLKTKVER 1283 +KA+ R K+ +R L+ +E R+LQR+LE+ ET +A ++E+ LK K+ R Sbjct: 1865 ADKASTRLKQLKRQLEEAEEEAQRANASRRKLQRELEDATETADAMNREVSSLKNKLRR 1923
Score = 73.9 bits (180), Expect = 1e-12
Identities = 92/460 (20%), Positives = 177/460 (38%), Gaps = 27/460 (5%)
Frame = +3
Query: 9 KSLLNRQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKERI 188
K + + + EL ++ L+ K+ E L+ ++ +E++N L K+ +
Sbjct: 938 KKKMQQNIQELEEQLEEEESARQKLQLEKVTTEAKLKKLEEDQIIMEDQNCKLAKEKKLL 997
Query: 189 EMQLTATKAQLDRELSSKDEMHDEQFKILNKK---LRDTEATLEEEQKQRTSLASLKKKL 359
E ++ A+ L ++E K+ NK + D E L E+KQR L ++KL
Sbjct: 998 EDRV----AEFTTNLMEEEEKSKSLAKLKNKHEAMITDLEERLRREEKQRQELEKTRRKL 1053
Query: 360 EIENVDLMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRL 539
E ++ DL +Q Q+ K + + ++E KN ALK+I+ELE ++
Sbjct: 1054 EGDSTDLSDQIAELQAQIAELKMQLAKKEEELQAALARVEEEAAQKNMALKKIRELETQI 1113
Query: 540 RAAESSTNQLIEDLSQSERIKR-------TLISERDDALEEIAT-----------LTVNK 665
+ +++E+ KR L +E +D L+ A +++ K
Sbjct: 1114 SELQEDLESERASRNKAEKQKRDLGEELEALKTELEDTLDSTAAQQELRSKREQEVSILK 1173
Query: 666 DTAIADKKRIESNLSALNMDYXXXXXXXXXXXDKNKKVTNQLEQVQNELNVER----QNA 833
T + K E+ + + + ++ K+V LE+ + L ER
Sbjct: 1174 KTLEDEAKTHEAQIQEMRQKHSQAVEELADQLEQTKRVKATLEKAKQTLENERGELANEV 1233
Query: 834 TRLENQKSTAE--RLNXXXXXXXXXXXXXTGKKYKATVSAQQIKIQSLDAQLDAKIEEIN 1007
L K +E R G++ + ++ K+ L +LD+ ++
Sbjct: 1234 KALLQGKGDSEHKRKKVEAQLQELQVKFSEGERVRTELAD---KVTKLQVELDSVTGLLS 1290
Query: 1008 QAXXXXXXXXXXXXXIMALMVDEQRRADNAIEQIEKATGRYKKAERSLQMEQENNSALTT 1187
Q+ + + + D Q Q + + K QME E NS
Sbjct: 1291 QSDSKSSKLTKDFSALESQLQDTQELLQEENRQKLSLSTKLK------QMEDEKNS---- 1340
Query: 1188 QNRRLQRDLEEVNETKEARDQEIKVLKTKVERLERRVRTG 1307
+ LEE E K +++I L +V +++++ G
Sbjct: 1341 ----FREQLEEEEEAKRNLEKQIATLHAQVTDMKKKMEDG 1376
Score = 67.8 bits (164), Expect = 8e-11
Identities = 83/415 (20%), Positives = 166/415 (40%), Gaps = 14/415 (3%)
Frame = +3
Query: 81 LKNAKIQLENDLENCKKTIEELEEENATLQMNKERIEMQLTATKAQLDRELSSKDEMHDE 260
L+ + +LE D + I EL+ + A L+M + E +L A A+++ E + K+
Sbjct: 1046 LEKTRRKLEGDSTDLSDQIAELQAQIAELKMQLAKKEEELQAALARVEEEAAQKN----- 1100
Query: 261 QFKILNKKLRDTEATLEEEQKQRTSLASLKKKLEIENVDLMNQXXXXXXXXXXXXRQIKK 440
+ KK+R+ E + E Q+ S + + K E + DL +
Sbjct: 1101 ---MALKKIRELETQISELQEDLESERASRNKAEKQKRDLGEELEALKTELEDTLDSTAA 1157
Query: 441 HQGTSLNSQREMQEVLNAKNEALKQIKELEKRLRAAESSTNQLIEDLSQS---------- 590
Q L S+RE QEV K + K E +++ +Q +E+L+
Sbjct: 1158 QQ--ELRSKRE-QEVSILKKTLEDEAKTHEAQIQEMRQKHSQAVEELADQLEQTKRVKAT 1214
Query: 591 -ERIKRTLISERDDALEEIATLTVNKDTAIADKKRIESNLSALNMDYXXXXXXXXXXXDK 767
E+ K+TL +ER + E+ L K + +K++E+ L L + + DK
Sbjct: 1215 LEKAKQTLENERGELANEVKALLQGKGDSEHKRKKVEAQLQELQVKFSEGERVRTELADK 1274
Query: 768 NKKVTNQLEQVQNELNVERQNATRLENQKSTAERLNXXXXXXXXXXXXXTGKKYKATVSA 947
K+ +L+ V L+ +++L S E + K ++S
Sbjct: 1275 VTKLQVELDSVTGLLSQSDSKSSKLTKDFSALES-----QLQDTQELLQEENRQKLSLST 1329
Query: 948 QQIKIQSLDAQLDAKIEEINQAXXXXXXXXXXXXXIMALMVDEQRRADNAIEQIEKATGR 1127
K++ ++ + ++ E++ + + A + D +++ ++ + +E A
Sbjct: 1330 ---KLKQMEDEKNSFREQLEEEEEAKRNLEKQIATLHAQVTDMKKKMEDGVGCLETAE-- 1384
Query: 1128 YKKAERSLQMEQENNSALTTQNRRLQRDLEEVN---ETKEARDQEIKVLKTKVER 1283
RRLQ+DLE ++ E K A +++ KT++++
Sbjct: 1385 -------------------EAKRRLQKDLEGLSQRLEEKVAAYDKLEKTKTRLQQ 1420
Score = 65.1 bits (157), Expect = 5e-10
Identities = 83/485 (17%), Positives = 184/485 (37%), Gaps = 46/485 (9%)
Frame = +3
Query: 6 SKSLLNRQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKER 185
+K L + EL + + + G ++ + ++E L+ + E E L +
Sbjct: 1218 AKQTLENERGELANEVKALLQGKGDSEHKRKKVEAQLQELQVKFSEGERVRTELADKVTK 1277
Query: 186 IEMQLTATKAQLDRELSSKDEMHDEQFKILNKKLRDTEATLEEEQKQRTSLASLKKKLEI 365
++++L + L + SK + F L +L+DT+ L+EE +Q+ SL++ K++E
Sbjct: 1278 LQVELDSVTGLLSQS-DSKSSKLTKDFSALESQLQDTQELLQEENRQKLSLSTKLKQMED 1336
Query: 366 ENVDLMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRLRA 545
E Q +QI + +++M++ + A + + L+K L
Sbjct: 1337 EKNSFREQLEEEEEAKRNLEKQIATLHAQVTDMKKKMEDGVGCLETAEEAKRRLQKDLEG 1396
Query: 546 AESSTNQLIEDLSQSERIKRTLISERDDALEEI--------------------------- 644
+ + + E+ K L E DD L ++
Sbjct: 1397 LSQRLEEKVAAYDKLEKTKTRLQQELDDLLVDLDHQRQSVSNLEKKQKKFDQLLAEEKTI 1456
Query: 645 -ATLTVNKDTAIADKKRIESNLSALNMDYXXXXXXXXXXXDKNKKVTNQLEQVQNELNVE 821
A +D A A+ + E+ +L NK+ ++E + + +
Sbjct: 1457 SAKYAEERDRAEAEAREKETKALSLARALEEAMEQKAELERLNKQFRTEMEDLMSSKDDV 1516
Query: 822 RQNATRLENQKSTAERLNXXXXXXXXXXXXXTGKKYKATVSAQ---QIKIQSLDAQLDAK 992
++ LE K E+ + +AT A+ ++ +Q++ AQ +
Sbjct: 1517 GKSVHELEKSKRALEQ----QVEEMKTQLEELEDELQATEDAKLRLEVNLQAMKAQFERD 1572
Query: 993 IE-EINQAXXXXXXXXXXXXXIMALMVDEQRRADNAIEQIEKATGRYKKAERSLQMEQEN 1169
++ Q+ + A + DE+++ A+ +K K E + +N
Sbjct: 1573 LQGRDEQSEEKKKQLVRQVREMEAELEDERKQRSMAMAARKKLEMDLKDLEAHIDTANKN 1632
Query: 1170 NSALTTQNRRLQRDLE----EVNETKEARDQ----------EIKVLKTKVERLERRVRTG 1307
Q R+LQ ++ E+++T+ +R++ ++K ++ ++ +L+ +
Sbjct: 1633 REEAIKQLRKLQAQMKDCMRELDDTRASREEILAQAKENEKKLKSMEAEMIQLQEELAAA 1692
Query: 1308 GRANR 1322
RA R
Sbjct: 1693 ERAKR 1697
Score = 53.5 bits (127), Expect = 2e-06
Identities = 84/454 (18%), Positives = 162/454 (35%), Gaps = 49/454 (10%)
Frame = +3
Query: 72 IGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKERIEMQLTATKAQLDRELSSKDEM 251
+ S+++ L + E K + L EN +M E ++ QL A K QL +L ++ E+
Sbjct: 838 LNSIRHEDELLAKEAELTKVREKHLAAENRLTEM--ETMQSQLMAEKLQLQEQLQAETEL 895
Query: 252 HDEQFKI----------LNKKLRDTEATLEEEQKQRTSLASLK----------------- 350
E ++ L + D EA +EEE+++ L + K
Sbjct: 896 CAEAEELRARLTAKKQELEEICHDLEARVEEEEERCQYLQAEKKKMQQNIQELEEQLEEE 955
Query: 351 ------------------KKLEIENVDLMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREM 476
KKLE + + + +Q ++ + + + +
Sbjct: 956 ESARQKLQLEKVTTEAKLKKLEEDQIIMEDQNCKLAKEKKLLEDRVAEFTTNLMEEEEKS 1015
Query: 477 QEVLNAKNEALKQIKELEKRLRAAESSTNQLIEDLSQSERIKRTLISERDDALEEIATLT 656
+ + KN+ I +LE+RLR E +L E+ +R L + D ++IA L
Sbjct: 1016 KSLAKLKNKHEAMITDLEERLRREEKQRQEL-------EKTRRKLEGDSTDLSDQIAELQ 1068
Query: 657 VNKDTAIADKKRIESNLSALNMDYXXXXXXXXXXXDKNKKVTNQLEQVQNELNVERQNAT 836
+ E L A K +++ Q+ ++Q +L ER +
Sbjct: 1069 AQIAELKMQLAKKEEELQAALARVEEEAAQKNMALKKIRELETQISELQEDLESERASRN 1128
Query: 837 RLENQK-STAERLNXXXXXXXXXXXXXTGKKYKATVSAQQIKIQSLDAQLDAKIEEINQA 1013
+ E QK E L ++ + Q++ I + +AK E
Sbjct: 1129 KAEKQKRDLGEELEALKTELEDTLDSTAAQQELRSKREQEVSILKKTLEDEAKTHEAQ-- 1186
Query: 1014 XXXXXXXXXXXXXIMALMVDEQRRADNAIEQIEKATGRYKKAERSLQMEQENNSALTTQN 1193
I + + + +Q+E+ E++ Q + L +
Sbjct: 1187 -------------IQEMRQKHSQAVEELADQLEQTKRVKATLEKAKQTLENERGELANEV 1233
Query: 1194 RRL---QRDLEEVNETKEARDQEIKVLKTKVERL 1286
+ L + D E + EA+ QE++V ++ ER+
Sbjct: 1234 KALLQGKGDSEHKRKKVEAQLQELQVKFSEGERV 1267
Score = 38.1 bits (87), Expect = 0.070
Identities = 31/178 (17%), Positives = 75/178 (42%), Gaps = 1/178 (0%)
Frame = +3
Query: 21 NRQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEE-NATLQMNKERIEMQ 197
N Q++++ ++ + + +NA+ QLE + K ++E+E + + + +E +
Sbjct: 1756 NLQIDQINTDLNLERSHAQKNENARQQLERQNKELKAKLQEMESAVKSKYKASIAALEAK 1815
Query: 198 LTATKAQLDRELSSKDEMHDEQFKILNKKLRDTEATLEEEQKQRTSLASLKKKLEIENVD 377
+ + QLD E + + +Q + KKL+D +E+E++ K
Sbjct: 1816 IAQLEEQLDNETKER-QAASKQVRRTEKKLKDVLLQVEDERRNAEQFKDQADKASTRLKQ 1874
Query: 378 LMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRLRAAE 551
L Q + ++ + QRE+++ + +++ L+ +LR +
Sbjct: 1875 LKRQ-------LEEAEEEAQRANASRRKLQRELEDATETADAMNREVSSLKNKLRRGD 1925
>sp|Q99105|MYSU_RABIT Myosin heavy chain, embryonic smooth muscle isoform Length = 501 Score = 238 bits (607), Expect = 4e-62 Identities = 137/422 (32%), Positives = 224/422 (53%) Frame = +3 Query: 18 LNRQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKERIEMQ 197 L + +L+S+KDDVGKN+ L+ +K LE +E + +EELE+E + K R+E+ Sbjct: 34 LRADMEDLMSSKDDVGKNVHELEKSKRALEQQVEEMRTQLEELEDELQATEDAKLRLEVN 93 Query: 198 LTATKAQLDRELSSKDEMHDEQFKILNKKLRDTEATLEEEQKQRTSLASLKKKLEIENVD 377 A KAQ +R+L ++DE +E+ ++L K++R+ EA LE+E+KQR + KKK+EI+ D Sbjct: 94 TQAMKAQFERDLQARDEQSEEKKRLLTKQVRELEAELEDERKQRALAVASKKKMEIDLKD 153 Query: 378 LMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRLRAAESS 557 L Q +Q+++ Q + QRE++E +++E Q KE EK+L++ E+ Sbjct: 154 LEAQIEAANKARERRVKQLRRLQAQMKDYQRELEEARGSRDEIFAQSKESEKKLKSLEAE 213 Query: 558 TNQLIEDLSQSERIKRTLISERDDALEEIATLTVNKDTAIADKKRIESNLSALNMDYXXX 737 QL E+L+ SER +R ERD+ +EIA K + +K+R+E+ + L + Sbjct: 214 ILQLQEELASSERARRHAEQERDELADEIANSASGKSALLDEKRRLEARMRQLEEELEEE 273 Query: 738 XXXXXXXXDKNKKVTNQLEQVQNELNVERQNATRLENQKSTAERLNXXXXXXXXXXXXXT 917 D+ +K T Q++ + EL ER A + +N + ER N Sbjct: 274 QSNMELLNDRFRKTTLQVDTLNAELAAERSAAQKSDNARQQLERQNKDLKAKLQELEGAV 333 Query: 918 GKKYKATVSAQQIKIQSLDAQLDAKIEEINQAXXXXXXXXXXXXXIMALMVDEQRRADNA 1097 K+KAT+SA + KI L+ QL+ + +E A I + DE+R AD Sbjct: 334 KSKFKATISALEAKIGQLEEQLEQEAKERAAANKLVRRTEKKLKEIFMQVEDERRHADQY 393 Query: 1098 IEQIEKATGRYKKAERSLQMEQENNSALTTQNRRLQRDLEEVNETKEARDQEIKVLKTKV 1277 EQ+EKA R K+ +R L+ +E + R+LQR+L++ E E +E+ LK ++ Sbjct: 394 KEQMEKANARMKQLKRQLEEAEEEATRANASRRKLQRELDDATEANEGLSREVSTLKNRL 453 Query: 1278 ER 1283 R Sbjct: 454 RR 455
Score = 50.4 bits (119), Expect = 1e-05
Identities = 55/306 (17%), Positives = 127/306 (41%), Gaps = 13/306 (4%)
Frame = +3
Query: 426 RQIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRLRAAESSTNQLIEDLSQSERIKR 605
R+ ++ + +L+ R ++E L AK E +Q K+L + SS + + +++ + E+ KR
Sbjct: 1 REAREKETKALSLSRALEEALEAKEEFERQNKQLRADMEDLMSSKDDVGKNVHELEKSKR 60
Query: 606 TLISERDDALEEIATLTVNKDTAIADKKRIESNLSALNMDYXXXXXXXXXXXDKNKKV-T 782
L + ++ ++ L K R+E N A+ + ++ K++ T
Sbjct: 61 ALEQQVEEMRTQLEELEDELQATEDAKLRLEVNTQAMKAQFERDLQARDEQSEEKKRLLT 120
Query: 783 NQLEQVQNELNVER-QNATRLENQKSTAERLNXXXXXXXXXXXXXTGKKYKATVSAQQIK 959
Q+ +++ EL ER Q A + ++K L + +A A++ +
Sbjct: 121 KQVRELEAELEDERKQRALAVASKKKMEIDLKDLEA------------QIEAANKARERR 168
Query: 960 IQSLDAQLDAKIEEINQAXXXXXXXXXXXXXIMALMVDEQRRADNAIEQIEKATGRYKKA 1139
++ L +L A++++ + + + + I Q+++ ++A
Sbjct: 169 VKQL-RRLQAQMKDYQRELEEARGSRDEIFAQSKESEKKLKSLEAEILQLQEELASSERA 227
Query: 1140 ERSLQMEQEN-----------NSALTTQNRRLQRDLEEVNETKEARDQEIKVLKTKVERL 1286
R + E++ SAL + RRL+ + ++ E E +++L + +
Sbjct: 228 RRHAEQERDELADEIANSASGKSALLDEKRRLEARMRQLEEELEEEQSNMELLNDRFRKT 287
Query: 1287 ERRVRT 1304
+V T
Sbjct: 288 TLQVDT 293
>sp|P14105|MYH9_CHICK Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A) Length = 1959 Score = 235 bits (599), Expect = 3e-61 Identities = 138/419 (32%), Positives = 229/419 (54%) Frame = +3 Query: 27 QLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKERIEMQLTA 206 ++ +L+S+KDDVGK++ L+ AK LE +E K +EELE+E + K R+E+ A Sbjct: 1505 EMEDLMSSKDDVGKSVHELEKAKRALEQQVEEMKTQLEELEDELQATEDAKLRLEVNQQA 1564 Query: 207 TKAQLDRELSSKDEMHDEQFKILNKKLRDTEATLEEEQKQRTSLASLKKKLEIENVDLMN 386 KAQ DR+L +DE ++E+ K L +++R+ E LE+E+KQR+ + +KKLE++ DL + Sbjct: 1565 MKAQFDRDLLGRDEQNEEKRKQLIRQVREMEVELEDERKQRSIAVAARKKLELDLKDLES 1624 Query: 387 QXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRLRAAESSTNQ 566 + ++K Q + RE+++ ++ E L Q KE EK+L++ E+ Q Sbjct: 1625 HIDTANKNRDEAIKHVRKLQAQMKDYMRELEDTRTSREEILAQAKENEKKLKSMEAEMIQ 1684 Query: 567 LIEDLSQSERIKRTLISERDDALEEIATLTVNKDTAIADKKRIESNLSALNMDYXXXXXX 746 L E+L+ +ER KR ERD+ +EIA + A+ +K+R+E+ ++ L + Sbjct: 1685 LQEELAAAERAKRQAQQERDELADEIANSSGKGALAMEEKRRLEARIAQLEEELEEEQGN 1744 Query: 747 XXXXXDKNKKVTNQLEQVQNELNVERQNATRLENQKSTAERLNXXXXXXXXXXXXXTGKK 926 D+ KK Q++Q+ +LN ER NA + EN + ER N K Sbjct: 1745 TEIINDRLKKANLQIDQMNADLNAERSNAQKNENARQQMERQNKELKLKLQEMESAVKSK 1804 Query: 927 YKATVSAQQIKIQSLDAQLDAKIEEINQAXXXXXXXXXXXXXIMALMVDEQRRADNAIEQ 1106 YKAT++A + KI L+ QLD + +E A I+ + DE+R A+ +Q Sbjct: 1805 YKATITALEAKIVQLEEQLDMETKERQAASKQVRRAEKKLKDILLQVDDERRNAEQFKDQ 1864 Query: 1107 IEKATGRYKKAERSLQMEQENNSALTTQNRRLQRDLEEVNETKEARDQEIKVLKTKVER 1283 +KA R K+ +R L+ E E + R+LQR+L++ ET +A ++E+ LK+K+ R Sbjct: 1865 ADKANMRLKQLKRQLE-EAEEEAQRANVRRKLQRELDDATETADAMNREVSSLKSKLRR 1922
Score = 78.2 bits (191), Expect = 6e-14
Identities = 94/460 (20%), Positives = 180/460 (39%), Gaps = 27/460 (5%)
Frame = +3
Query: 9 KSLLNRQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKERI 188
K + + + EL ++ L+ K+ E L+ ++ + LE++N L K+ +
Sbjct: 938 KKKMQQNIQELEEQLEEEESARQKLQLEKVTTEAKLKKLEEDVIVLEDQNLKLAKEKKLL 997
Query: 189 EMQLTATKAQLDRELSSKDEMHDEQFKILNKK---LRDTEATLEEEQKQRTSLASLKKKL 359
E +++ + L+ ++E K+ NK + D E L E+KQR L ++KL
Sbjct: 998 EDRMS----EFTTNLTEEEEKSKSLAKLKNKHEAMITDLEERLRREEKQRQELEKTRRKL 1053
Query: 360 EIENVDLMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRL 539
E ++ DL +Q Q+ K + + ++E KN ALK+I+ELE ++
Sbjct: 1054 EGDSSDLHDQIAELQAQIAELKIQLSKKEEELQAALARVEEEAAQKNMALKKIRELESQI 1113
Query: 540 RAAESSTNQLIEDLSQSERIKR-------TLISERDDALEEIAT-----------LTVNK 665
+ +++E+ KR L +E +D L+ A +TV K
Sbjct: 1114 TELQEDLESERASRNKAEKQKRDLGEELEALKTELEDTLDSTAAQQELRSKREQEVTVLK 1173
Query: 666 DTAIADKKRIESNLSALNMDYXXXXXXXXXXXDKNKKVTNQLEQVQNELNVERQNATR-- 839
T + K E+ + + + ++ K+V LE+ + L ER +
Sbjct: 1174 KTLEDEAKTHEAQIQEMRQKHSQAIEELAEQLEQTKRVKANLEKAKQALESERAELSNEV 1233
Query: 840 --LENQKSTAE--RLNXXXXXXXXXXXXXTGKKYKATVSAQQIKIQSLDAQLDAKIEEIN 1007
L K AE R G++ K ++ ++ L +LD +N
Sbjct: 1234 KVLLQGKGDAEHKRKKVDAQLQELQVKFTEGERVKTELAE---RVNKLQVELDNVTGLLN 1290
Query: 1008 QAXXXXXXXXXXXXXIMALMVDEQRRADNAIEQIEKATGRYKKAERSLQMEQENNSALTT 1187
Q+ + + + D Q E +++ T L+ ++ +AL
Sbjct: 1291 QSDSKSIKLAKDFSALESQLQDTQ-------ELLQEETRLKLSFSTKLKQTEDEKNALKE 1343
Query: 1188 QNRRLQRDLEEVNETKEARDQEIKVLKTKVERLERRVRTG 1307
Q LEE E K +++I VL+ + +++ G
Sbjct: 1344 Q-------LEEEEEAKRNLEKQISVLQQQAVEARKKMDDG 1376
Score = 55.1 bits (131), Expect = 6e-07
Identities = 82/439 (18%), Positives = 170/439 (38%), Gaps = 22/439 (5%)
Frame = +3
Query: 9 KSLLNRQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKERI 188
K+ L + L S + ++ + L K E+ + ++EL+ + + K +
Sbjct: 1212 KANLEKAKQALESERAELSNEVKVLLQGKGDAEHKRKKVDAQLQELQVKFTEGERVKTEL 1271
Query: 189 EMQLTATKAQLDR------ELSSKDEMHDEQFKILNKKLRDTEATLEEEQKQRTSLASLK 350
++ + +LD + SK + F L +L+DT+ L+EE + + S ++
Sbjct: 1272 AERVNKLQVELDNVTGLLNQSDSKSIKLAKDFSALESQLQDTQELLQEETRLKLSFSTKL 1331
Query: 351 KKLEIENVDLMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVLNAKNEALKQIKELE 530
K+ E E L Q +QI Q ++ ++++M + L A + K+L+
Sbjct: 1332 KQTEDEKNALKEQLEEEEEAKRNLEKQISVLQQQAVEARKKMDDGLGCLEIAEEAKKKLQ 1391
Query: 531 KRLRAAESSTNQLIEDLSQSERIKRTLISERDDALEEIATLTVNKDTAIADKKRIESNLS 710
K L + + I + E+ K L E DD ++ +K+ + L+
Sbjct: 1392 KDLESLTQRYEEKIAAYDKLEKTKTRLQQELDDIAVDLDHQRQTVSNLEKKQKKFDQLLA 1451
Query: 711 -ALNMDYXXXXXXXXXXXDKNKKVTNQL-------EQVQNELNVERQN---ATRLENQKS 857
N+ + +K T L E ++ + +ER N T +E+ S
Sbjct: 1452 EEKNISAKYAEERDRAEAEAREKETKALSLARALEEAIEQKAELERVNKQFRTEMEDLMS 1511
Query: 858 TAERLNXXXXXXXXXXXXXTGKKYKATVSAQQIKIQSLDAQLDAKIE-EINQAXXXXXXX 1034
+ + + + + ++++ L A DAK+ E+NQ
Sbjct: 1512 SKDDVGKSVHELEKAKRALEQQVEEMKTQLEELE-DELQATEDAKLRLEVNQQAMKAQFD 1570
Query: 1035 XXXXXXIMALMVDEQRRADNAIEQIEKATGRYKKAERSLQMEQENNSALTTQNRRLQRDL 1214
L DEQ E+ ++ + ++ E L+ E++ S ++L+ DL
Sbjct: 1571 RD------LLGRDEQNE-----EKRKQLIRQVREMEVELEDERKQRSIAVAARKKLELDL 1619
Query: 1215 EE----VNETKEARDQEIK 1259
++ ++ + RD+ IK
Sbjct: 1620 KDLESHIDTANKNRDEAIK 1638
Score = 52.8 bits (125), Expect = 3e-06
Identities = 68/306 (22%), Positives = 128/306 (41%), Gaps = 20/306 (6%)
Frame = +3
Query: 18 LNRQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEEL-----EEENATLQMNKE 182
L ++ N+D+ K++ L+ +LE+ + + EE+ E E M E
Sbjct: 1622 LESHIDTANKNRDEAIKHVRKLQAQMKDYMRELEDTRTSREEILAQAKENEKKLKSMEAE 1681
Query: 183 RIEMQ--LTATKAQLDRELSSKDEMHDEQFKILNKKLRDTEATLEEEQKQRTSLASLKKK 356
I++Q L A + + +DE+ DE I N + A +EE+++ +A L+++
Sbjct: 1682 MIQLQEELAAAERAKRQAQQERDELADE---IANSSGKGALA-MEEKRRLEARIAQLEEE 1737
Query: 357 LEIE--NVDLMNQXXXXXXXXXXXXRQIKKHQGTSLNSQR-EMQEVLNAKNEALKQIKEL 527
LE E N +++N QI + LN++R Q+ NA+ + +Q KEL
Sbjct: 1738 LEEEQGNTEIINDRLKKANL------QIDQ-MNADLNAERSNAQKNENARQQMERQNKEL 1790
Query: 528 EKRLRAAESSTNQLIEDLSQSERIKRTLISERDDALEEIATLTVNKDTAIADKKRIESNL 707
+ +L+ ES+ + + K + E+ D T + A +R E L
Sbjct: 1791 KLKLQEMESAVKSKYKATITALEAKIVQLEEQLD------METKERQAASKQVRRAEKKL 1844
Query: 708 SALNMDYXXXXXXXXXXXDKNKKVTNQLEQVQNEL----------NVERQNATRLENQKS 857
+ + D+ K +L+Q++ +L NV R+ L++
Sbjct: 1845 KDILLQVDDERRNAEQFKDQADKANMRLKQLKRQLEEAEEEAQRANVRRKLQRELDDATE 1904
Query: 858 TAERLN 875
TA+ +N
Sbjct: 1905 TADAMN 1910
Score = 52.0 bits (123), Expect = 5e-06
Identities = 84/433 (19%), Positives = 159/433 (36%), Gaps = 49/433 (11%)
Frame = +3
Query: 138 EELEEENATLQMNKERIEMQLTATKAQLDRELSSKDEMHDEQFKI----------LNKKL 287
++L EN +M E + QL A K QL +L ++ E+ E +I L +
Sbjct: 860 KQLAAENRLSEM--ETFQAQLMAEKMQLQEQLQAEAELCAEAEEIRARLTAKKQELEEIC 917
Query: 288 RDTEATLEEEQKQRTSLASLKKKLEIENVDLMNQXXXXXXXXXXXXRQ------------ 431
D EA +EEE+++ L + KKK++ +L Q +
Sbjct: 918 HDLEARVEEEEERCQHLQAEKKKMQQNIQELEEQLEEEESARQKLQLEKVTTEAKLKKLE 977
Query: 432 ----IKKHQGTSLNSQREMQE-------------------VLNAKNEALKQIKELEKRLR 542
+ + Q L ++++ E + KN+ I +LE+RLR
Sbjct: 978 EDVIVLEDQNLKLAKEKKLLEDRMSEFTTNLTEEEEKSKSLAKLKNKHEAMITDLEERLR 1037
Query: 543 AAESSTNQLIEDLSQSERIKRTLISERDDALEEIATLTVNKDTAIADKKRIESNLSALNM 722
E +L E+ +R L + D ++IA L + E L A
Sbjct: 1038 REEKQRQEL-------EKTRRKLEGDSSDLHDQIAELQAQIAELKIQLSKKEEELQAALA 1090
Query: 723 DYXXXXXXXXXXXDKNKKVTNQLEQVQNELNVERQNATRLENQK-STAERLNXXXXXXXX 899
K +++ +Q+ ++Q +L ER + + E QK E L
Sbjct: 1091 RVEEEAAQKNMALKKIRELESQITELQEDLESERASRNKAEKQKRDLGEELEALKTELED 1150
Query: 900 XXXXXTGKKYKATVSAQQIKIQSLDAQLDAKIEEINQAXXXXXXXXXXXXXIMALMVDEQ 1079
++ + Q++ + + +AK E I +
Sbjct: 1151 TLDSTAAQQELRSKREQEVTVLKKTLEDEAKTHEAQ---------------IQEMRQKHS 1195
Query: 1080 RRADNAIEQIE---KATGRYKKAERSLQMEQENNSALTTQNRRLQRDLEEVNETKEARDQ 1250
+ + EQ+E + +KA+++L+ E+ S + + D E + +A+ Q
Sbjct: 1196 QAIEELAEQLEQTKRVKANLEKAKQALESERAELSNEVKVLLQGKGDAEHKRKKVDAQLQ 1255
Query: 1251 EIKVLKTKVERLE 1289
E++V T+ ER++
Sbjct: 1256 ELQVKFTEGERVK 1268
Score = 35.4 bits (80), Expect = 0.45
Identities = 30/178 (16%), Positives = 72/178 (40%), Gaps = 1/178 (0%)
Frame = +3
Query: 21 NRQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEE-NATLQMNKERIEMQ 197
N Q++++ ++ + N +NA+ Q+E + K ++E+E + + +E +
Sbjct: 1756 NLQIDQMNADLNAERSNAQKNENARQQMERQNKELKLKLQEMESAVKSKYKATITALEAK 1815
Query: 198 LTATKAQLDRELSSKDEMHDEQFKILNKKLRDTEATLEEEQKQRTSLASLKKKLEIENVD 377
+ + QLD E + + +Q + KKL+D +++E++ K +
Sbjct: 1816 IVQLEEQLDMETKER-QAASKQVRRAEKKLKDILLQVDDERRNAEQFKDQADKANMRLKQ 1874
Query: 378 LMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRLRAAE 551
L Q + + QRE+ + + +++ L+ +LR +
Sbjct: 1875 LKRQ--------LEEAEEEAQRANVRRKLQRELDDATETADAMNREVSSLKSKLRRGD 1924
Score = 31.2 bits (69), Expect = 8.5
Identities = 25/104 (24%), Positives = 47/104 (45%), Gaps = 5/104 (4%)
Frame = +3
Query: 24 RQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEE--NATLQMNKERIEMQ 197
++L +++ DD +N K+ + L+ K+ +EE EEE A ++ +R
Sbjct: 1842 KKLKDILLQVDDERRNAEQFKDQADKANMRLKQLKRQLEEAEEEAQRANVRRKLQRELDD 1901
Query: 198 LTATKAQLDRELS---SKDEMHDEQFKILNKKLRDTEATLEEEQ 320
T T ++RE+S SK D F + + +R +E+
Sbjct: 1902 ATETADAMNREVSSLKSKLRRGDLPFVVTRRLVRKGTGECSDEE 1945
>sp|Q62812|MYH9_RAT Myosin-9 (Myosin heavy chain, nonmuscle IIa) (Nonmuscle myosin heavy chain IIa) (NMMHC II-a) (NMMHC-IIA) (Cellular myosin heavy chain, type A) (Nonmuscle myosin heavy chain-A) (NMMHC-A) Length = 1961 Score = 233 bits (594), Expect = 1e-60 Identities = 134/419 (31%), Positives = 228/419 (54%) Frame = +3 Query: 27 QLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKERIEMQLTA 206 ++ +L+S+KDDVGK++ L+ + LE +E K +EELE+E + K R+E+ L A Sbjct: 1506 EMEDLMSSKDDVGKSVHELEKSNRALEQQVEEMKTQLEELEDELQATEDAKLRLEVNLQA 1565 Query: 207 TKAQLDRELSSKDEMHDEQFKILNKKLRDTEATLEEEQKQRTSLASLKKKLEIENVDLMN 386 KAQ +R+L +DE +E+ K L +++R+ EA LE+E+KQR+ + +KKLE++ DL Sbjct: 1566 MKAQFERDLQGRDEQSEEKKKQLVRQVREMEAELEDERKQRSIAMAARKKLEMDLKDLEA 1625 Query: 387 QXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRLRAAESSTNQ 566 +Q++K Q + R++ + ++ E L Q KE EK+L++ E+ Q Sbjct: 1626 HIDTANKNREEAIKQLRKLQAQMKDCMRDVDDTRASREEILAQAKENEKKLKSMEAEMIQ 1685 Query: 567 LIEDLSQSERIKRTLISERDDALEEIATLTVNKDTAIADKKRIESNLSALNMDYXXXXXX 746 L E+L+ +ER KR ERD+ +EIA + A+ +K+R+E+ ++ L + Sbjct: 1686 LQEELAAAERAKRQAQQERDELADEIANSSGKGALALEEKRRLEALIALLEEELEEEQGN 1745 Query: 747 XXXXXDKNKKVTNQLEQVQNELNVERQNATRLENQKSTAERLNXXXXXXXXXXXXXTGKK 926 D+ KK Q++Q+ +LN+ER +A + EN + ER N K Sbjct: 1746 TELINDRLKKANLQIDQINTDLNLERSHAQKNENARQQLERQNKELKAKLQEMESAVKSK 1805 Query: 927 YKATVSAQQIKIQSLDAQLDAKIEEINQAXXXXXXXXXXXXXIMALMVDEQRRADNAIEQ 1106 YKA+++A + KI L+ QLD + +E A ++ + DE+R A+ +Q Sbjct: 1806 YKASIAALEAKIAQLEEQLDNETKERQAASKQVRRAEKKLKDVLLQVEDERRNAEQFKDQ 1865 Query: 1107 IEKATGRYKKAERSLQMEQENNSALTTQNRRLQRDLEEVNETKEARDQEIKVLKTKVER 1283 +KA+ R K+ +R L+ +E R+LQR+LE+ ET +A ++E+ LK K+ R Sbjct: 1866 ADKASTRLKQLKRQLEEAEEEAQRANASRRKLQRELEDATETADAMNREVSSLKNKLRR 1924
Score = 72.0 bits (175), Expect = 4e-12
Identities = 93/460 (20%), Positives = 179/460 (38%), Gaps = 27/460 (5%)
Frame = +3
Query: 9 KSLLNRQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKERI 188
K + + + EL ++ L+ K+ E L+ ++ +E++N L K+ +
Sbjct: 938 KKKMQQNIQELEEQLEEEESARQKLQLEKVTTEAKLKKLEEDQIIMEDQNCKLAKEKKLL 997
Query: 189 EMQLTATKAQLDRELSSKDEMHDEQFKILNKK---LRDTEATLEEEQKQRTSLASLKKKL 359
E ++ A+ +L ++E K+ NK + D E L E+KQR L ++KL
Sbjct: 998 EDRV----AEFTTDLMEEEEKSKSLAKLKNKHEAMITDLEERLRREEKQRQELEKTRRKL 1053
Query: 360 EIENVDLMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRL 539
E ++ DL +Q Q+ K + + ++E KN ALK+I+ELE ++
Sbjct: 1054 EGDSTDLSDQIAELQAQIAELKMQLAKKEEELQAALARVEEEAAQKNMALKKIRELETQI 1113
Query: 540 RAAESSTNQLIEDLSQSERIKR-------TLISERDDALEEIAT-----------LTVNK 665
+ +++E+ KR L +E +D L+ A +++ K
Sbjct: 1114 SELQEDLESERACRNKAEKQKRDLGEELEALKTELEDTLDSTAAQQELRSKREQEVSILK 1173
Query: 666 DTAIADKKRIESNLSALNMDYXXXXXXXXXXXDKNKKVTNQLEQVQNELNVER----QNA 833
T + K E+ + + + ++ K+V LE+ + L ER
Sbjct: 1174 KTLEDEAKTHEAQIQEMRQKHSQAVEELAEQLEQTKRVKATLEKAKQTLENERGELANEV 1233
Query: 834 TRLENQKSTAE--RLNXXXXXXXXXXXXXTGKKYKATVSAQQIKIQSLDAQLDAKIEEIN 1007
L K +E R G++ + ++ K+ L +LD+ +N
Sbjct: 1234 KALLQGKGDSEHKRKKVEAQLQELQVKFSEGERVRTELAD---KVSKLQVELDSVTGLLN 1290
Query: 1008 QAXXXXXXXXXXXXXIMALMVDEQRRADNAIEQIEKATGRYKKAERSLQMEQENNSALTT 1187
Q+ + + + D Q Q + + K QME E NS
Sbjct: 1291 QSDSKSSKLTKDFSALESQLQDTQELLQEENRQKLSLSTKLK------QMEDEKNSF--- 1341
Query: 1188 QNRRLQRDLEEVNETKEARDQEIKVLKTKVERLERRVRTG 1307
+L+ EE E K +++I L +V +++++ G
Sbjct: 1342 -REQLE---EEEEEAKRNLEKQIATLHAQVTDMKKKMEDG 1377
Score = 69.7 bits (169), Expect = 2e-11
Identities = 85/415 (20%), Positives = 167/415 (40%), Gaps = 14/415 (3%)
Frame = +3
Query: 81 LKNAKIQLENDLENCKKTIEELEEENATLQMNKERIEMQLTATKAQLDRELSSKDEMHDE 260
L+ + +LE D + I EL+ + A L+M + E +L A A+++ E + K+
Sbjct: 1046 LEKTRRKLEGDSTDLSDQIAELQAQIAELKMQLAKKEEELQAALARVEEEAAQKN----- 1100
Query: 261 QFKILNKKLRDTEATLEEEQKQRTSLASLKKKLEIENVDLMNQXXXXXXXXXXXXRQIKK 440
+ KK+R+ E + E Q+ S + + K E + DL +
Sbjct: 1101 ---MALKKIRELETQISELQEDLESERACRNKAEKQKRDLGEELEALKTELEDTLDSTAA 1157
Query: 441 HQGTSLNSQREMQEVLNAKNEALKQIKELEKRLRAAESSTNQLIEDLSQS---------- 590
Q L S+RE QEV K + K E +++ +Q +E+L++
Sbjct: 1158 QQ--ELRSKRE-QEVSILKKTLEDEAKTHEAQIQEMRQKHSQAVEELAEQLEQTKRVKAT 1214
Query: 591 -ERIKRTLISERDDALEEIATLTVNKDTAIADKKRIESNLSALNMDYXXXXXXXXXXXDK 767
E+ K+TL +ER + E+ L K + +K++E+ L L + + DK
Sbjct: 1215 LEKAKQTLENERGELANEVKALLQGKGDSEHKRKKVEAQLQELQVKFSEGERVRTELADK 1274
Query: 768 NKKVTNQLEQVQNELNVERQNATRLENQKSTAERLNXXXXXXXXXXXXXTGKKYKATVSA 947
K+ +L+ V LN +++L S E + K ++S
Sbjct: 1275 VSKLQVELDSVTGLLNQSDSKSSKLTKDFSALES-----QLQDTQELLQEENRQKLSLST 1329
Query: 948 QQIKIQSLDAQLDAKIEEINQAXXXXXXXXXXXXXIMALMVDEQRRADNAIEQIEKATGR 1127
K++ ++ + ++ E++ + + V + ++ ++E G
Sbjct: 1330 ---KLKQMEDEKNSFREQLEEEEEEAKRNLEKQIATLHAQVTDMKK------KMEDGVGC 1380
Query: 1128 YKKAERSLQMEQENNSALTTQNRRLQRDLEEVN---ETKEARDQEIKVLKTKVER 1283
+ AE + RRLQ+DLE ++ E K A +++ KT++++
Sbjct: 1381 LETAEEA--------------KRRLQKDLEGLSQRLEEKVAAYDKLEKTKTRLQQ 1421
Score = 68.2 bits (165), Expect = 6e-11
Identities = 101/489 (20%), Positives = 192/489 (39%), Gaps = 55/489 (11%)
Frame = +3
Query: 18 LNRQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKERIEMQ 197
L+ Q+ ++ +D + + + AK +L+ DLE + +EE L+ K R++ +
Sbjct: 1363 LHAQVTDMKKKMEDGVGCLETAEEAKRRLQKDLEGLSQRLEEKVAAYDKLEKTKTRLQQE 1422
Query: 198 LTATKAQLDRELSSKDEMHDEQFKI----------------------LNKKLRDTEA--- 302
L LD + S + +Q K + ++T+A
Sbjct: 1423 LDDLLVDLDHQRQSVSNLEKKQKKFDQLLAEEKTISAKYAEERDRAEAEAREKETKALSL 1482
Query: 303 --TLEEEQKQRTSLASLKKKLEIENVDLMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREM 476
LEE +Q+ L L K+ E DLM+ K G S++ +
Sbjct: 1483 ARALEEAMEQKAELERLNKQFRTEMEDLMSS---------------KDDVGKSVHELEKS 1527
Query: 477 QEVLNAKNEALK-QIKELEKRLRAAESSTNQLIEDL---------------SQSERIKRT 608
L + E +K Q++ELE L+A E + +L +L QSE K+
Sbjct: 1528 NRALEQQVEEMKTQLEELEDELQATEDAKLRLEVNLQAMKAQFERDLQGRDEQSEEKKKQ 1587
Query: 609 LISERDDALEEIATLTVNKDTAIADKKRIESNLSALNMDYXXXXXXXXXXXDKNKKVTNQ 788
L+ + + E+ + A+A +K++E +L L + +K+ Q
Sbjct: 1588 LVRQVREMEAELEDERKQRSIAMAARKKLEMDLKDLEAHIDTANKNREEAIKQLRKLQAQ 1647
Query: 789 LEQVQNELNVERQNATRLENQKSTAERLNXXXXXXXXXXXXXTGKKYKATVSAQQIKIQS 968
++ +++ R + + Q E KK K ++ A+ I++Q
Sbjct: 1648 MKDCMRDVDDTRASREEILAQAKENE------------------KKLK-SMEAEMIQLQE 1688
Query: 969 LDAQLDAKIEEINQAXXXXXXXXXXXXXIMALMVDEQRRADNAI----EQIEKATG---- 1124
A + + Q AL ++E+RR + I E++E+ G
Sbjct: 1689 ELAAAERAKRQAQQERDELADEIANSSGKGALALEEKRRLEALIALLEEELEEEQGNTEL 1748
Query: 1125 ---RYKKAERSLQMEQENNSALTTQNRRLQRDLEEVNE-TKEARDQEIKVLKTKVERLER 1292
R KKA +LQ++Q N + L+R + NE ++ +++ K LK K++ +E
Sbjct: 1749 INDRLKKA--NLQIDQINT------DLNLERSHAQKNENARQQLERQNKELKAKLQEMES 1800
Query: 1293 RVRTGGRAN 1319
V++ +A+
Sbjct: 1801 AVKSKYKAS 1809
Score = 54.3 bits (129), Expect = 9e-07
Identities = 86/454 (18%), Positives = 161/454 (35%), Gaps = 49/454 (10%)
Frame = +3
Query: 72 IGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKERIEMQLTATKAQLDRELSSKDEM 251
+ S+++ L + E K + L EN +M E ++ QL A K QL +L +K E+
Sbjct: 838 LNSIRHEDELLAKEAELTKVREKHLAAENRLTEM--ETMQSQLMAEKLQLQEQLQAKTEL 895
Query: 252 HDEQFKI----------LNKKLRDTEATLEEEQKQRTSLASLK----------------- 350
E ++ L + D EA +EEE+++ L + K
Sbjct: 896 CAEAEELRARLTAKKQELEEICHDLEARVEEEEERCQYLQAEKKKMQQNIQELEEQLEEE 955
Query: 351 ------------------KKLEIENVDLMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREM 476
KKLE + + + +Q ++ + + + +
Sbjct: 956 ESARQKLQLEKVTTEAKLKKLEEDQIIMEDQNCKLAKEKKLLEDRVAEFTTDLMEEEEKS 1015
Query: 477 QEVLNAKNEALKQIKELEKRLRAAESSTNQLIEDLSQSERIKRTLISERDDALEEIATLT 656
+ + KN+ I +LE+RLR E +L E+ +R L + D ++IA L
Sbjct: 1016 KSLAKLKNKHEAMITDLEERLRREEKQRQEL-------EKTRRKLEGDSTDLSDQIAELQ 1068
Query: 657 VNKDTAIADKKRIESNLSALNMDYXXXXXXXXXXXDKNKKVTNQLEQVQNELNVERQNAT 836
+ E L A K +++ Q+ ++Q +L ER
Sbjct: 1069 AQIAELKMQLAKKEEELQAALARVEEEAAQKNMALKKIRELETQISELQEDLESERACRN 1128
Query: 837 RLENQK-STAERLNXXXXXXXXXXXXXTGKKYKATVSAQQIKIQSLDAQLDAKIEEINQA 1013
+ E QK E L ++ + Q++ I + +AK E
Sbjct: 1129 KAEKQKRDLGEELEALKTELEDTLDSTAAQQELRSKREQEVSILKKTLEDEAKTHEAQ-- 1186
Query: 1014 XXXXXXXXXXXXXIMALMVDEQRRADNAIEQIEKATGRYKKAERSLQMEQENNSALTTQN 1193
I + + + EQ+E+ E++ Q + L +
Sbjct: 1187 -------------IQEMRQKHSQAVEELAEQLEQTKRVKATLEKAKQTLENERGELANEV 1233
Query: 1194 RRL---QRDLEEVNETKEARDQEIKVLKTKVERL 1286
+ L + D E + EA+ QE++V ++ ER+
Sbjct: 1234 KALLQGKGDSEHKRKKVEAQLQELQVKFSEGERV 1267
Score = 38.1 bits (87), Expect = 0.070
Identities = 31/178 (17%), Positives = 75/178 (42%), Gaps = 1/178 (0%)
Frame = +3
Query: 21 NRQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEE-NATLQMNKERIEMQ 197
N Q++++ ++ + + +NA+ QLE + K ++E+E + + + +E +
Sbjct: 1757 NLQIDQINTDLNLERSHAQKNENARQQLERQNKELKAKLQEMESAVKSKYKASIAALEAK 1816
Query: 198 LTATKAQLDRELSSKDEMHDEQFKILNKKLRDTEATLEEEQKQRTSLASLKKKLEIENVD 377
+ + QLD E + + +Q + KKL+D +E+E++ K
Sbjct: 1817 IAQLEEQLDNETKER-QAASKQVRRAEKKLKDVLLQVEDERRNAEQFKDQADKASTRLKQ 1875
Query: 378 LMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRLRAAE 551
L Q + ++ + QRE+++ + +++ L+ +LR +
Sbjct: 1876 LKRQ-------LEEAEEEAQRANASRRKLQRELEDATETADAMNREVSSLKNKLRRGD 1926
Score = 32.7 bits (73), Expect = 2.9
Identities = 23/105 (21%), Positives = 49/105 (46%), Gaps = 6/105 (5%)
Frame = +3
Query: 24 RQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKERIEMQL- 200
++L +++ +D +N K+ + L+ K+ +EE EEE ++ +++ +L
Sbjct: 1843 KKLKDVLLQVEDERRNAEQFKDQADKASTRLKQLKRQLEEAEEEAQRANASRRKLQRELE 1902
Query: 201 --TATKAQLDRELSS---KDEMHDEQFKILNKKLRDTEATLEEEQ 320
T T ++RE+SS K D F + + +R +E+
Sbjct: 1903 DATETADAMNREVSSLKNKLRRGDMPFVVTRRIVRKGTGDCSDEE 1947
>sp|P35749|MYH11_HUMAN Myosin-11 (Myosin heavy chain, smooth muscle isoform) (SMMHC) Length = 1972 Score = 232 bits (591), Expect = 3e-60 Identities = 131/426 (30%), Positives = 228/426 (53%) Frame = +3 Query: 6 SKSLLNRQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKER 185 + +L ++ +LVS+KDDVGKN+ L+ +K LE +E K +EELE+E + K R Sbjct: 1505 TNKMLKAEMEDLVSSKDDVGKNVHELEKSKRALETQMEEMKTQLEELEDELQATEDAKLR 1564 Query: 186 IEMQLTATKAQLDRELSSKDEMHDEQFKILNKKLRDTEATLEEEQKQRTSLASLKKKLEI 365 +E+ + A K Q +R+L ++DE ++E+ + L ++L + E LE+E+KQR A+ KKKLE Sbjct: 1565 LEVNMQALKGQFERDLQARDEQNEEKRRQLQRQLHEYETELEDERKQRALAAAAKKKLEG 1624 Query: 366 ENVDLMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRLRA 545 + DL Q +Q++K Q + QRE+++ +++E KE EK+ ++ Sbjct: 1625 DLKDLELQADSAIKGREEAIKQLRKLQAQMKDFQRELEDARASRDEIFATAKENEKKAKS 1684 Query: 546 AESSTNQLIEDLSQSERIKRTLISERDDALEEIATLTVNKDTAIADKKRIESNLSALNMD 725 E+ QL EDL+ +ER ++ E+++ EE+A+ ++ +K+R+E+ ++ L + Sbjct: 1685 LEADLMQLQEDLAAAERARKQADLEKEELAEELASSLSGRNALQDEKRRLEARIAQLEEE 1744 Query: 726 YXXXXXXXXXXXDKNKKVTNQLEQVQNELNVERQNATRLENQKSTAERLNXXXXXXXXXX 905 D+ +K T Q EQ+ NEL ER A + E+ + ER N Sbjct: 1745 LEEEQGNMEAMSDRVRKATQQAEQLSNELATERSTAQKNESARQQLERQNKELRSKLHEM 1804 Query: 906 XXXTGKKYKATVSAQQIKIQSLDAQLDAKIEEINQAXXXXXXXXXXXXXIMALMVDEQRR 1085 K+K+T++A + KI L+ Q++ + E A I+ + DE++ Sbjct: 1805 EGAVKSKFKSTIAALEAKIAQLEEQVEQEAREKQAATKSLKQKDKKLKEILLQVEDERKM 1864 Query: 1086 ADNAIEQIEKATGRYKKAERSLQMEQENNSALTTQNRRLQRDLEEVNETKEARDQEIKVL 1265 A+ EQ EK R K+ +R L+ +E + + R+LQR+L+E E+ EA +E+ L Sbjct: 1865 AEQYKEQAEKGNARVKQLKRQLEEAEEESQRINANRRKLQRELDEATESNEAMGREVNAL 1924 Query: 1266 KTKVER 1283 K+K+ R Sbjct: 1925 KSKLRR 1930
Score = 72.0 bits (175), Expect = 4e-12
Identities = 97/487 (19%), Positives = 187/487 (38%), Gaps = 35/487 (7%)
Frame = +3
Query: 18 LNRQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKERIEMQ 197
+ RQ E+ + +D++ K K + + EN+L+ ++ +L EE LQ + + E +
Sbjct: 847 VTRQEEEMQAKEDELQKT----KERQQKAENELKELEQKHSQLTEEKNLLQ-EQLQAETE 901
Query: 198 LTATKAQLDRELSSKDEMHDEQFKILNKKLRDTEATLEEEQKQRTSLASLKKKLEIENVD 377
L A ++ L++K + L + L + EA LEEE+ + L + +KK+ + +D
Sbjct: 902 LYAEAEEMRVRLAAKKQE-------LEEILHEMEARLEEEEDRGQQLQAERKKMAQQMLD 954
Query: 378 LMNQ-------XXXXXXXXXXXXRQIKK---------HQGTSLNSQREMQE--------- 482
L Q +IKK Q L+ +R++ E
Sbjct: 955 LEEQLEEEEAARQKLQLEKVTAEAKIKKLEDEILVMDDQNNKLSKERKLLEERISDLTTN 1014
Query: 483 ----------VLNAKNEALKQIKELEKRLRAAESSTNQLIEDLSQSERIKRTLISERDDA 632
+ KN+ I ELE RL+ E S +L E++KR L + D
Sbjct: 1015 LAEEEEKAKNLTKLKNKHESMISELEVRLKKEEKSRQEL-------EKLKRKLEGDASDF 1067
Query: 633 LEEIATLTVNKDTAIADKKRIESNLSALNMDYXXXXXXXXXXXDKNKKVTNQLEQVQNEL 812
E+IA L + E L A K +++ + +Q +L
Sbjct: 1068 HEQIADLQAQIAELKMQLAKKEEELQAALARLDDEIAQKNNALKKIRELEGHISDLQEDL 1127
Query: 813 NVERQNATRLENQKSTAERLNXXXXXXXXXXXXXTGKKYKATVSAQQIKIQSLDAQLDAK 992
+ ER + E QK G++ +A + + + S Q + +
Sbjct: 1128 DSERAARNKAEKQK------------------RDLGEELEALKTELEDTLDSTATQQELR 1169
Query: 993 IEEINQAXXXXXXXXXXXXXIMALMVDEQRRADNAIEQIEKATGRYKKAERSLQMEQENN 1172
+ + A + + +++ A+E++ + ++K+A+ +L +N
Sbjct: 1170 AKREQEVTVLKKALDEETRSHEAQVQEMRQKHAQAVEELTEQLEQFKRAKANL---DKNK 1226
Query: 1173 SALTTQNRRLQRDLEEVNETKEARDQEIKVLKTKVERLERRVRTGGRANRXXXXXXXXXX 1352
L +N L +L + + K+ + + K L+ +V+ L+ + G RA
Sbjct: 1227 QTLEKENADLAGELRVLGQAKQEVEHKKKKLEAQVQELQSKCSDGERARAELNDKVHKLQ 1286
Query: 1353 XXXESIT 1373
ES+T
Sbjct: 1287 NEVESVT 1293
Score = 68.2 bits (165), Expect = 6e-11
Identities = 102/497 (20%), Positives = 190/497 (38%), Gaps = 62/497 (12%)
Frame = +3
Query: 9 KSLLNRQLNELVSNKDDVGKNIGSLKNAKIQLENDLENCKKTIEELEEENATLQMNKERI 188
++ L QL+E + K ++ ++I +L + L++ T+E LEE Q E +
Sbjct: 1345 RNSLQDQLDEEMEAKQNLERHISTLNIQLSDSKKKLQDFASTVEALEEGKKRFQKEIENL 1404
Query: 189 EMQ----------LTATKAQLDRELSSKDEMHDEQFKI---LNKKLRDTEATLEEEQKQR 329
Q L TK +L +EL D Q ++ L KK R + L EE+
Sbjct: 1405 TQQYEEKAAAYDKLEKTKNRLQQELDDLVVDLDNQRQLVSNLEKKQRKFDQLLAEEKNIS 1464
Query: 330 TSLASLKKKLEIENVDLMNQXXXXXXXXXXXXRQIKKHQGTSLNSQREMQEVLNAKNEAL 509
+ A + + E E + + ++ + T+ + EM++++++K++
Sbjct: 1465 SKYADERDRAEAEAREKETKALSLARALEEALEAKEELERTNKMLKAEMEDLVSSKDDVG 1524
Query: 510 KQIKELEKRLRAAESSTNQL---IEDL--------------------------------- 581
K + ELEK RA E+ ++ +E+L
Sbjct: 1525 KNVHELEKSKRALETQMEEMKTQLEELEDELQATEDAKLRLEVNMQALKGQFERDLQARD 1584
Query: 582 SQSERIKRTLISERDDALEEIATLTVNKDTAIADKKRIESNLSALNMDYXXXXXXXXXXX 761
Q+E +R L + + E+ + A A KK++E +L L +
Sbjct: 1585 EQNEEKRRQLQRQLHEYETELEDERKQRALAAAAKKKLEGDLKDLELQADSAIKGREEAI 1644
Query: 762 DKNKKVTNQLEQVQNELNVERQN-----ATRLENQKSTAERLNXXXXXXXXXXXXXTGKK 926
+ +K+ Q++ Q EL R + AT EN+K A+ L +
Sbjct: 1645 KQLRKLQAQMKDFQRELEDARASRDEIFATAKENEKK-AKSLEADLMQLQEDLAAAERAR 1703
Query: 927 YKATVSAQQIKIQSLDAQLDAKIEEINQAXXXXXXXXXXXXXIMALMVDEQRRADNAIEQ 1106
+A + ++ L +L + + N + + +EQ + ++
Sbjct: 1704 KQADLEKEE-----LAEELASSLSGRNALQDEKRRLEARIAQLEEELEEEQGNMEAMSDR 1758
Query: 1107 IEKATGRYKKAERSLQME----QENNSA---LTTQNRRLQRDLEEV-NETKEARDQEIKV 1262
+ KAT + ++ L E Q+N SA L QN+ L+ L E+ K I
Sbjct: 1759 VRKATQQAEQLSNELATERSTAQKNESARQQLERQNKELRSKLHEMEGAVKSKFKSTIAA 1818
Query: 1263 LKTKVERLERRVRTGGR 1313
L+ K+ +LE +V R
Sbjct: 1819 LEAKIAQLEEQVEQEAR 1835
Score = 65.9 bits (159), Expect = 3e-10
Identities = 88/426 (20%), Positives = 172/426 (40%), Gaps = 22/426 (5%)
Frame = +3
Query: 93 KIQLENDLENCKKTIEELEEENATL--QMNKERIEMQLTATK-AQLDRELSSKDEMHDEQ 263
K+QLE K I++LE+E + Q NK E +L + + L L+ ++E
Sbjct: 968 KLQLEKVTAEAK--IKKLEDEILVMDDQNNKLSKERKLLEERISDLTTNLAEEEEKAKNL 1025
Query: 264 FKILNKK---LRDTEATLEEEQKQRTSLASLKKKLEIENVDLMNQXXXXXXXXXXXXRQI 434
K+ NK + + E L++E+K R L LK+KLE + D Q Q+
Sbjct: 1026 TKLKNKHESMISELEVRLKKEEKSRQELEKLKRKLEGDASDFHEQIADLQAQIAELKMQL 1085
Query: 435 KKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRLRAAESSTNQLIEDLSQSERIKR--- 605
K + + + + + KN ALK+I+ELE + + + +++E+ KR
Sbjct: 1086 AKKEEELQAALARLDDEIAQKNNALKKIRELEGHISDLQEDLDSERAARNKAEKQKRDLG 1145
Query: 606 ----TLISERDDALEEIATLTVNKDTAIADKKRIESNLSALNMDYXXXXXXXXXXXDKNK 773
L +E +D L+ AT + K+ E + +D + +
Sbjct: 1146 EELEALKTELEDTLDSTAT-----QQELRAKREQEVTVLKKALD-EETRSHEAQVQEMRQ 1199
Query: 774 KVTNQLEQVQNELNVERQNATRLENQKSTAERLN----XXXXXXXXXXXXXTGKKYKATV 941
K +E++ +L ++ L+ K T E+ N KK K
Sbjct: 1200 KHAQAVEELTEQLEQFKRAKANLDKNKQTLEKENADLAGELRVLGQAKQEVEHKKKKLEA 1259
Query: 942 SAQQIKIQSLD-----AQLDAKIEEINQAXXXXXXXXXXXXXIMALMVDEQRRADNAIEQ 1106
Q+++ + D A+L+ K+ ++ + ++ + + +A +
Sbjct: 1260 QVQELQSKCSDGERARAELNDKVHKLQN----------EVESVTGMLNEAEGKAIKLAKD 1309
Query: 1107 IEKATGRYKKAERSLQMEQENNSALTTQNRRLQRDLEEVNETKEARDQEIKVLKTKVERL 1286
+ + + + + LQ E ++T+ R+L+ EE N ++ D+E++ + L
Sbjct: 1310 VASLSSQLQDTQELLQEETRQKLNVSTKLRQLE---EERNSLQDQLDEEMEA----KQNL 1362
Query: 1287 ERRVRT 1304
ER + T
Sbjct: 1363 ERHIST 1368
Score = 40.4 bits (93), Expect = 0.014
Identities = 49/291 (16%), Positives = 107/291 (36%)
Frame = +3
Query: 429 QIKKHQGTSLNSQREMQEVLNAKNEALKQIKELEKRLRAAESSTNQLIEDLSQSERIKRT 608
Q+ + + + E+Q+ + +A ++KELE++ N L E L + +
Sbjct: 846 QVTRQEEEMQAKEDELQKTKERQQKAENELKELEQKHSQLTEEKNLLQEQL----QAETE 901
Query: 609 LISERDDALEEIATLTVNKDTAIADKKRIESNLSALNMDYXXXXXXXXXXXDKNKKVTNQ 788
L +E ++ +A A K+ +E L + + KK+ Q
Sbjct: 902 LYAEAEEMRVRLA----------AKKQELEEILHEMEARLEEEEDRGQQLQAERKKMAQQ 951
Query: 789 LEQVQNELNVERQNATRLENQKSTAERLNXXXXXXXXXXXXXTGKKYKATVSAQQIKIQS 968
+ ++ +L E +L+ +K TAE KI+
Sbjct: 952 MLDLEEQLEEEEAARQKLQLEKVTAE-----------------------------AKIKK 982
Query: 969 LDAQLDAKIEEINQAXXXXXXXXXXXXXIMALMVDEQRRADNAIEQIEKATGRYKKAERS 1148
L+ ++ ++ N+ + + +E+ +A N + K + E
Sbjct: 983 LEDEILVMDDQNNKLSKERKLLEERISDLTTNLAEEEEKAKNLTKLKNKHESMISELEVR 1042
Query: 1149 LQMEQENNSALTTQNRRLQRDLEEVNETKEARDQEIKVLKTKVERLERRVR 1301
L+ E+++ L R+L+ D + +E +I LK ++ + E ++
Sbjct: 1043 LKKEEKSRQELEKLKRKLEGDASDFHEQIADLQAQIAELKMQLAKKEEELQ 1093
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 129,789,226
Number of Sequences: 369166
Number of extensions: 2141608
Number of successful extensions: 12358
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 8793
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 11100
length of database: 68,354,980
effective HSP length: 115
effective length of database: 47,110,455
effective search space used: 18797071545
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail