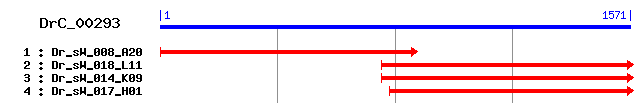
DrC_00293
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00293 (1571 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P04068|HYEP_RABIT Epoxide hydrolase 1 (Microsomal epoxid... 325 2e-88 sp|P07099|HYEP_HUMAN Epoxide hydrolase 1 (Microsomal epoxid... 324 5e-88 sp|P07687|HYEP_RAT Epoxide hydrolase 1 (Microsomal epoxide ... 320 6e-87 sp|Q9D379|HYEP_MOUSE Epoxide hydrolase 1 (Microsomal epoxid... 318 3e-86 sp|Q8MZR5|HYEP2_CTEFE Juvenile hormone epoxide hydrolase 2 ... 305 2e-82 sp|Q8MZR6|HYEP1_CTEFE Juvenile hormone epoxide hydrolase 1 ... 302 2e-81 sp|Q25489|HYEP_MANSE Juvenile hormone epoxide hydrolase (Ju... 254 6e-67 sp|Q06816|HYEP_STIAU Epoxide hydrolase (Epoxide hydratase) 70 1e-11 sp|O52866|HYES_CORS2 Soluble epoxide hydrolase (SEH) (Epoxi... 42 0.006 sp|P53750|YN93_YEAST Hypothetical 32.8 kDa protein in BIO3-... 41 0.008
>sp|P04068|HYEP_RABIT Epoxide hydrolase 1 (Microsomal epoxide hydrolase) (Epoxide hydratase) Length = 455 Score = 325 bits (833), Expect = 2e-88 Identities = 176/431 (40%), Positives = 261/431 (60%), Gaps = 12/431 (2%) Frame = +3 Query: 126 KDSW---GSR--NINDKSITEILIKVPIETLSNLFARIKNRVSAPSFSNSDFQYGFNVEY 290 +D W GSR + D+SI ++ E +++L RI P NS F YGFN Y Sbjct: 30 EDGWWGPGSRPVGLEDESIRPFKVETSDEEINDLHQRIDRIRLTPPLENSRFHYGFNSNY 89 Query: 291 LNKMMDYWLNKYDWQHWEEVINRFPHRFTIIKGLQIHYIHV-XXXXXXXXXXXXXXXXHG 467 L K++ YW +++DW+ E++N +PH T I+GL IH+IHV HG Sbjct: 90 LKKILSYWRHEFDWKKQVEILNSYPHFKTKIEGLDIHFIHVKPPQVPPGRTPKPLLMVHG 149 Query: 468 WPGSFFEFYKLIPLLSTQTE---KDQYAFEIVCPSLVGYGWSDGATIPGFDLFAMSDVMF 638 WPGSFFEFYK+IPLL+ D++ FE++CPS+ GYG+S ++ GF+ + + + + Sbjct: 150 WPGSFFEFYKIIPLLTDPKSHGLSDEHIFEVICPSIPGYGFSQASSKKGFNSVSTARIFY 209 Query: 639 TLMERLNITSKFYIQGGDFGFIIARSMSVINPKKVAGIHINFVETPYHDMRYLTQVFIGS 818 LM RL +FYIQGGD+G ++ +M+ + P V G+H+N + Y + +G Sbjct: 210 KLMLRLGF-QEFYIQGGDWGALVCTNMAQLVPSHVKGLHLNMALILRN--HYTLTLLLGR 266 Query: 819 MLPAKLHFLTQRELDMVYPWSDKI-NTILSSMGYFGLQSTRPSTIGIALSDSPAGLAAYI 995 + L + T+R+++++YP+ +K+ +++ GY +++T+P T+G AL+DSP GLAAYI Sbjct: 267 RIGGLLGY-TERDMELLYPFKEKVFYSLMRESGYMHIRATKPDTVGCALNDSPVGLAAYI 325 Query: 996 LEKFGAWSNNNSYHLSSPNGGLEDIKITKDEMLTNVMIYYITNTISSSVRLYKEMLNE-- 1169 LEKF W+N+ L +GGLE K + ++LTN+MIY+ T +I SS R YKE L + Sbjct: 326 LEKFSTWTNSEFRDLE--DGGLER-KFSLQDLLTNIMIYWTTGSIVSSQRYYKENLGQGF 382 Query: 1170 QNHKLFKTPVRVPTAIAMFPNEIIQVPKPFVHFKYRNVRQFNYMPRGGHFAALEEPELLA 1349 HK + V VPT A FP EI+ VP+ +V KY + ++YMPRGGHFAA EEPELLA Sbjct: 383 MAHKHERLKVHVPTGFAAFPCEIMHVPEKWVRTKYPQLISYSYMPRGGHFAAFEEPELLA 442 Query: 1350 NDIRSFINLME 1382 DI F+ L+E Sbjct: 443 RDICKFVGLVE 453
>sp|P07099|HYEP_HUMAN Epoxide hydrolase 1 (Microsomal epoxide hydrolase) (Epoxide hydratase) Length = 455 Score = 324 bits (830), Expect = 5e-88 Identities = 180/435 (41%), Positives = 262/435 (60%), Gaps = 16/435 (3%) Frame = +3 Query: 126 KDSW---GSRNI--NDKSITEILIKVPIETLSNLFARIKNRVSAPSFSNSDFQYGFNVEY 290 +D W G+R+ D SI ++ E + +L RI P +S F YGFN Y Sbjct: 30 EDGWWGPGTRSAAREDDSIRPFKVETSDEEIHDLHQRIDKFRFTPPLEDSCFHYGFNSNY 89 Query: 291 LNKMMDYWLNKYDWQHWEEVINRFPHRFTIIKGLQIHYIHV-XXXXXXXXXXXXXXXXHG 467 L K++ YW N++DW+ E++NR+PH T I+GL IH+IHV HG Sbjct: 90 LKKVISYWRNEFDWKKQVEILNRYPHFKTKIEGLDIHFIHVKPPQLPAGHTPKPLLMVHG 149 Query: 468 WPGSFFEFYKLIPLLSTQTE---KDQYAFEIVCPSLVGYGWSDGATIPGFDLFAMSDVMF 638 WPGSF+EFYK+IPLL+ D++ FE++CPS+ GYG+S+ ++ GF+ A + + + Sbjct: 150 WPGSFYEFYKIIPLLTDPKNHGLSDEHVFEVICPSIPGYGFSEASSKKGFNSVATARIFY 209 Query: 639 TLMERLNITSKFYIQGGDFGFIIARSMSVINPKKVAGIHIN--FVETPYHDMRYLTQVFI 812 LM RL +FYIQGGD+G +I +M+ + P V G+H+N V + + + L Sbjct: 210 KLMLRLGF-QEFYIQGGDWGSLICTNMAQLVPSHVKGLHLNMALVLSNFSTLTLLLGQRF 268 Query: 813 GSMLPAKLHFLTQRELDMVYPWSDKI-NTILSSMGYFGLQSTRPSTIGIALSDSPAGLAA 989 G L LT+R+++++YP +K+ +++ GY +Q T+P T+G AL+DSP GLAA Sbjct: 269 GRFLG-----LTERDVELLYPVKEKVFYSLMRESGYMHIQCTKPDTVGSALNDSPVGLAA 323 Query: 990 YILEKFGAWSNNNSYHLSSPNGGLEDIKITKDEMLTNVMIYYITNTISSSVRLYKEMLNE 1169 YILEKF W+N +L +GGLE K + D++LTNVM+Y+ T TI SS R YKE L + Sbjct: 324 YILEKFSTWTNTEFRYLE--DGGLER-KFSLDDLLTNVMLYWTTGTIISSQRFYKENLGQ 380 Query: 1170 ----QNHKLFKTPVRVPTAIAMFPNEIIQVPKPFVHFKYRNVRQFNYMPRGGHFAALEEP 1337 Q H+ K V VPT + FP E++ P+ +V FKY + ++YM RGGHFAA EEP Sbjct: 381 GWMTQKHERMK--VYVPTGFSAFPFELLHTPEKWVRFKYPKLISYSYMVRGGHFAAFEEP 438 Query: 1338 ELLANDIRSFINLME 1382 ELLA DIR F++++E Sbjct: 439 ELLAQDIRKFLSVLE 453
>sp|P07687|HYEP_RAT Epoxide hydrolase 1 (Microsomal epoxide hydrolase) (Epoxide hydratase) Length = 455 Score = 320 bits (821), Expect = 6e-87 Identities = 170/418 (40%), Positives = 253/418 (60%), Gaps = 9/418 (2%) Frame = +3 Query: 156 DKSITEILIKVPIETLSNLFARIKNRVSAPSFSNSDFQYGFNVEYLNKMMDYWLNKYDWQ 335 D+SI ++ E + +L RI ++P S F YGFN Y+ K++ YW N++DW+ Sbjct: 45 DESIRPFKVETSDEEIKDLHQRIDRFRASPPLEGSRFHYGFNSNYMKKVVSYWRNEFDWR 104 Query: 336 HWEEVINRFPHRFTIIKGLQIHYIHV-XXXXXXXXXXXXXXXXHGWPGSFFEFYKLIPLL 512 E++N++PH T I+GL IH+IHV HGWPGSF+EFYK+IPLL Sbjct: 105 KQVEILNQYPHFKTKIEGLDIHFIHVKPPQLPSGRTPKPLLMVHGWPGSFYEFYKIIPLL 164 Query: 513 STQTE---KDQYAFEIVCPSLVGYGWSDGATIPGFDLFAMSDVMFTLMERLNITSKFYIQ 683 + D++ FE++CPS+ GYG+S+ ++ G + A + + + LM RL KFYIQ Sbjct: 165 TDPKSHGLSDEHVFEVICPSIPGYGYSEASSKKGLNSVATARIFYKLMTRLGF-QKFYIQ 223 Query: 684 GGDFGFIIARSMSVINPKKVAGIHIN--FVETPYHDMRYLTQVFIGSMLPAKLHFLTQRE 857 GGD+G +I +M+ + P V G+H+N F+ ++ M L G L T+++ Sbjct: 224 GGDWGSLICTNMAQMVPNHVKGLHLNMAFISRSFYTMTPLLGQRFGRFLG-----YTEKD 278 Query: 858 LDMVYPWSDKI-NTILSSMGYFGLQSTRPSTIGIALSDSPAGLAAYILEKFGAWSNNNSY 1034 ++++YP+ +K+ +I+ GY +Q+T+P T+G AL+DSP GLAAYILEKF W+ + Sbjct: 279 IELLYPYKEKVFYSIMRESGYLHIQATKPDTVGCALNDSPVGLAAYILEKFSTWTKSEYR 338 Query: 1035 HLSSPNGGLEDIKITKDEMLTNVMIYYITNTISSSVRLYKEMLNE--QNHKLFKTPVRVP 1208 L +GGLE K + D++L N+MIY+ T TI SS R YKE L + HK V VP Sbjct: 339 ELE--DGGLER-KFSLDDLLVNIMIYWTTGTIVSSQRYYKENLGQGIMVHKHEGMKVFVP 395 Query: 1209 TAIAMFPNEIIQVPKPFVHFKYRNVRQFNYMPRGGHFAALEEPELLANDIRSFINLME 1382 T + FP+E++ P+ +V KY + ++YM RGGHFAA EEP+LLA DIR F++L E Sbjct: 396 TGFSAFPSELLHAPEKWVKVKYPKLISYSYMERGGHFAAFEEPKLLAQDIRKFVSLAE 453
>sp|Q9D379|HYEP_MOUSE Epoxide hydrolase 1 (Microsomal epoxide hydrolase) (Epoxide hydratase) Length = 455 Score = 318 bits (815), Expect = 3e-86 Identities = 171/418 (40%), Positives = 253/418 (60%), Gaps = 9/418 (2%) Frame = +3 Query: 156 DKSITEILIKVPIETLSNLFARIKNRVSAPSFSNSDFQYGFNVEYLNKMMDYWLNKYDWQ 335 D+SI ++ E + +L RI ++P S F YGFN YL K++ +W N++DW+ Sbjct: 45 DESIRPFKVETSDEEIKDLHQRIDRFRASPPLEGSRFHYGFNSSYLKKVVSFWRNEFDWR 104 Query: 336 HWEEVINRFPHRFTIIKGLQIHYIHV-XXXXXXXXXXXXXXXXHGWPGSFFEFYKLIPLL 512 E++N++PH T I+GL IH+IHV HGWPGSF+EFYK+IPLL Sbjct: 105 KQVEILNQYPHFKTKIEGLDIHFIHVKPPQLPSGRTPKPLLMVHGWPGSFYEFYKIIPLL 164 Query: 513 S---TQTEKDQYAFEIVCPSLVGYGWSDGATIPGFDLFAMSDVMFTLMERLNITSKFYIQ 683 + T D++ FE++CPS+ GYG+S+ ++ G + A + + + LM RL KFYIQ Sbjct: 165 TDPKTHGLSDEHVFEVICPSIPGYGFSEASSKKGLNSVATARIFYKLMSRLGF-QKFYIQ 223 Query: 684 GGDFGFIIARSMSVINPKKVAGIHIN--FVETPYHDMRYLTQVFIGSMLPAKLHFLTQRE 857 GGD+G +I +++ + P V G+H+N F+ + + L G L T+++ Sbjct: 224 GGDWGSLICTNIAQMVPNHVKGLHLNMSFISRNIYSLTPLLGQRFGRFLG-----YTEKD 278 Query: 858 LDMVYPWSDKI-NTILSSMGYFGLQSTRPSTIGIALSDSPAGLAAYILEKFGAWSNNNSY 1034 L+++YP+ +K+ I+ GY +Q+T+P T+G AL+DSP GLAAYILEKF W+ + Sbjct: 279 LELLYPFKEKVFYNIMRESGYLHIQATKPDTVGCALNDSPVGLAAYILEKFSTWTKSEYR 338 Query: 1035 HLSSPNGGLEDIKITKDEMLTNVMIYYITNTISSSVRLYKEMLNE--QNHKLFKTPVRVP 1208 L +GGLE K + +++LTN+MIY+ T TI SS R YKE L + H+ V VP Sbjct: 339 ELE--DGGLER-KFSLEDLLTNIMIYWTTGTIVSSQRFYKENLGQGVMVHRHEGMKVFVP 395 Query: 1209 TAIAMFPNEIIQVPKPFVHFKYRNVRQFNYMPRGGHFAALEEPELLANDIRSFINLME 1382 T + FP+EI+ PK +V KY + ++YM RGGHFAA EEP+LLA DIR F++L E Sbjct: 396 TGYSAFPSEILHAPKKWVKVKYPKLISYSYMERGGHFAAFEEPKLLAQDIRKFVSLAE 453
>sp|Q8MZR5|HYEP2_CTEFE Juvenile hormone epoxide hydrolase 2 (Juvenile hormone epoxide hydrolase II) (Juvenile hormone-specific epoxide hydrolase II) (JHEH II) (CfEH2) Length = 465 Score = 305 bits (782), Expect = 2e-82 Identities = 170/426 (39%), Positives = 246/426 (57%), Gaps = 6/426 (1%) Frame = +3 Query: 111 PNVTAKDSWGSRNIN--DKSITEILIKVPIETLSNLFARIKNRVSAPSFSNSDFQYGFNV 284 PN+ WG+ D S+ I + E L+ L ++ + P DFQYGFN Sbjct: 30 PNIPLDTWWGTGKSQKIDTSMRPFKIAINDEVLNTLKVKLSDVSFTPPLEGIDFQYGFNT 89 Query: 285 EYLNKMMDYWLNKYDWQHWEEVINRFPHRFTIIKGLQIHYIHVXXXXXXXXXXXXXXXXH 464 L K++D+W +Y+W+ E ++N++PH T I+GL IHY+H+ H Sbjct: 90 NTLKKLVDFWRTQYNWREREALLNKYPHFKTNIQGLDIHYVHIKPQVSKNIHVLPMIMVH 149 Query: 465 GWPGSFFEFYKLIPLLSTQTEKDQYAFEIVCPSLVGYGWSDGATIPGFDLFAMSDVMFTL 644 GWPGSF EFYK+IP+L+T + FE++ PS+ GYG+S A PG ++ +M L Sbjct: 150 GWPGSFVEFYKIIPMLTTPRTDYNFVFELILPSIPGYGFSQAAAKPGLGATQIAVIMHNL 209 Query: 645 MERLNITSKFYIQGGDFGFIIARSMSVINPKKVAGIHIN--FVETPYHDMRYLTQVFIGS 818 M+R+ K+Y+QGGD+G I +MS + P+ V G H N F+ T +++ F+GS Sbjct: 210 MDRIGF-KKYYVQGGDWGSRIVSAMSTLFPENVLGHHSNLCFLNTLSSNIK----SFVGS 264 Query: 819 MLPAKLHFLTQRELDMVYPWSDKINTILSSMGYFGLQSTRPSTIGIALSDSPAGLAAYIL 998 + P F ++ + +YP S+ T+L GYF +Q+T+P T+G+AL DSPAGLAAYIL Sbjct: 265 LFPE--WFAGKQNVHKIYPLSEHFFTLLEESGYFHIQATKPDTVGVALRDSPAGLAAYIL 322 Query: 999 EKFGAWSNNNSYHLSSPNGGLEDIKITKDEMLTNVMIYYITNTISSSVRLYKEMLNEQNH 1178 EKF + N S+ +G L+ K T E+L NVMIYY+T +I++S+R+Y E + + Sbjct: 323 EKFS--TGTNKAWRSAKDGNLQS-KFTFTELLDNVMIYYVTGSITTSMRIYAESYSWDHL 379 Query: 1179 KL--FKTPVRVPTAIAMFPNEIIQVPKPFVHFKYRNVRQFNYMPRGGHFAALEEPELLAN 1352 L + P VPTA A FP+EI + KY+ + Q MPRGGHFAALEEP LLA Sbjct: 380 SLNMDRVPTIVPTACAKFPHEIAYKTDFQLAEKYKTLLQSTIMPRGGHFAALEEPLLLAE 439 Query: 1353 DIRSFI 1370 DI S + Sbjct: 440 DIFSAV 445
>sp|Q8MZR6|HYEP1_CTEFE Juvenile hormone epoxide hydrolase 1 (Juvenile hormone epoxide hydrolase I) (Juvenile hormone-specific epoxide hydrolase I) (JHEH I) (CfEH1) Length = 464 Score = 302 bits (773), Expect = 2e-81 Identities = 171/423 (40%), Positives = 242/423 (57%), Gaps = 7/423 (1%) Frame = +3 Query: 111 PNVTAKDSWGS---RNINDKSITEILIKVPIETLSNLFARIKNRVSAPSFSNSDFQYGFN 281 PN+ WG +N+ D SI I + + + NL ++ + +F+YGFN Sbjct: 30 PNIPLDTWWGPGKPQNV-DISIRPFKININNKVIENLKLKLNDVQYTLPLEGINFEYGFN 88 Query: 282 VEYLNKMMDYWLNKYDWQHWEEVINRFPHRFTIIKGLQIHYIHVXXXXXXXXXXXXXXXX 461 + L K++D+W +Y+W+ E ++N++PH T I+GL IHY+H+ Sbjct: 89 TDSLKKIVDFWRTQYNWREREALLNKYPHFKTNIQGLDIHYVHIKPQVSKNIEVLPLVMI 148 Query: 462 HGWPGSFFEFYKLIPLLSTQTEKDQYAFEIVCPSLVGYGWSDGATIPGFDLFAMSDVMFT 641 HGWPGSF EFYK+IP+L+T + FE++ PS+ GYG+S A PG ++ +M Sbjct: 149 HGWPGSFVEFYKIIPMLTTPRAGYNFVFELILPSIPGYGFSQAAAKPGLGSTQVAVIMRN 208 Query: 642 LMERLNITSKFYIQGGDFGFIIARSMSVINPKKVAGIHIN--FVETPYHDMRYLTQVFIG 815 LMER+ K+Y+QGGD+G +I +MS + P+ V G H N FV TP +++ + IG Sbjct: 209 LMERIGF-KKYYVQGGDWGSMIISAMSTLFPENVLGQHSNMCFVNTPSSNIKAI----IG 263 Query: 816 SMLPAKLHFLTQRELDMVYPWSDKINTILSSMGYFGLQSTRPSTIGIALSDSPAGLAAYI 995 S P F +YP S+ T+L MGY LQ+T+P T+G+AL DSPAGLAAYI Sbjct: 264 SFFPES--FAGTGNAHKMYPMSEHFFTLLEEMGYLHLQATKPDTVGVALRDSPAGLAAYI 321 Query: 996 LEKFGAWSNNNSYHLSSPNGGLEDIKITKDEMLTNVMIYYITNTISSSVRLYKEMLNEQN 1175 LEKF W+N + S G +K E+L NVMIYY+T++I++S+RLY E + + Sbjct: 322 LEKFSTWTNRS---WRSVKDGNLLLKYNIPELLDNVMIYYVTDSITTSMRLYAESFTKAH 378 Query: 1176 HKLFKTPVR--VPTAIAMFPNEIIQVPKPFVHFKYRNVRQFNYMPRGGHFAALEEPELLA 1349 L VR VP A A FPNE+ V + KY+ + Q N MP GGHFAA EEP LLA Sbjct: 379 LALNLDRVRNHVPAACAKFPNELAYVTDCQLAEKYKTLLQSNDMPSGGHFAAFEEPGLLA 438 Query: 1350 NDI 1358 DI Sbjct: 439 EDI 441
>sp|Q25489|HYEP_MANSE Juvenile hormone epoxide hydrolase (Juvenile hormone-specific epoxide hydrolase) (JHEH) Length = 462 Score = 254 bits (648), Expect = 6e-67 Identities = 162/450 (36%), Positives = 241/450 (53%), Gaps = 9/450 (2%) Frame = +3 Query: 36 ISSFALIGGILLNSNCVADLFLSAVPNVTAKDS---WG---SRNINDKSITEILIKVPIE 197 +SSF + G+ + S V L VP D WG DKSI I Sbjct: 5 LSSF--VAGVAIGSGLVITYVLYNVPEPPELDLQRWWGIGTRPTEEDKSIRPFSIDFNDT 62 Query: 198 TLSNLFARIKNRVS-APSFSNSDFQYGFNVEYLNKMMDYWLNKYDWQHWEEVINRFPHRF 374 + +L R+KNR + +YG N EYL +++YWLN+Y+++ E++N+FPH Sbjct: 63 VILDLKERLKNRRPFTKPLEGINSEYGMNTEYLETVLEYWLNEYNFKKRAELLNKFPHYK 122 Query: 375 TIIKGLQIHYIHVXXXXXXXXXXXXXXXXHGWPGSFFEFYKLIPLLSTQTEKDQYAFEIV 554 T I+GL +H+I V HGWP S EF K+IP+L+T + FE+V Sbjct: 123 TRIQGLDLHFIRVKPEIKEGVQVLPLLMMHGWPSSSKEFDKVIPILTTPKHEYNIVFEVV 182 Query: 555 CPSLVGYGWSDGATIPGFDLFAMSDVMFTLMERLNITSKFYIQGGDFGFIIARSMSVINP 734 L GYG+S+G PG + + +M LM RL KFYIQ GD+G A M+ + P Sbjct: 183 AVDLPGYGFSEGTNKPGLNPVQIGVMMRNLMLRLGF-EKFYIQAGDWGSQCATHMATLFP 241 Query: 735 KKVAGIHINFVETPYHDMRYLT-QVFIGSMLPAKLHFLTQRELDMVYPWSDKINTILSSM 911 +V G+H N P T ++FIG++ P+ + + + +D +YP + + IL Sbjct: 242 DQVLGLHTNM---PLSSRPLSTVKLFIGALFPSLI--VDAKYMDRIYPLKNLFSYILRET 296 Query: 912 GYFGLQSTRPSTIGIALSDSPAGLAAYILEKFGAWSNNNSYHLSSPNGGLEDIKITKDEM 1091 GYF +Q+T+P TIG+AL+DSPAGLA Y++EK SN + L +P+GGLE++ + D++ Sbjct: 297 GYFHIQATKPDTIGVALTDSPAGLAGYLIEKMAICSNRD--QLDTPHGGLENLNL--DDV 352 Query: 1092 LTNVMIYYITNTISSSVRLYKEMLN-EQNHKLFKTPVRVPTAIAMFPNEIIQVPKPFVHF 1268 L V I +I N I +S RLY E + + + + P VPTA F E++ P + Sbjct: 353 LDTVTINWINNCIVTSTRLYAEGFSWPEVLIVHRIPSMVPTAGINFKYEVLYQPDWILRD 412 Query: 1269 KYRNVRQFNYMPRGGHFAALEEPELLANDI 1358 K+ N+ + + GGHFAAL P+ LA+DI Sbjct: 413 KFPNLVRSTVLDFGGHFAALHTPQALADDI 442
>sp|Q06816|HYEP_STIAU Epoxide hydrolase (Epoxide hydratase) Length = 232 Score = 70.5 bits (171), Expect = 1e-11 Identities = 36/98 (36%), Positives = 59/98 (60%), Gaps = 2/98 (2%) Frame = +3 Query: 1074 ITKDEMLTNVMIYYITNTISSSVRLYKEML--NEQNHKLFKTPVRVPTAIAMFPNEIIQV 1247 +++DEML N+ +Y++T+T +SS R+Y E N KL +P +++FP E+ + Sbjct: 131 LSQDEMLDNISLYWLTDTAASSARIYWENAGSNFSGGKL-----DLPVGVSVFpreLFRA 185 Query: 1248 PKPFVHFKYRNVRQFNYMPRGGHFAALEEPELLANDIR 1361 PK + Y + +N RGGHFAA E+P L A+++R Sbjct: 186 PKRWAEQTYSKLIYWNEPDRGGHFAAFEQPALFAHELR 223
>sp|O52866|HYES_CORS2 Soluble epoxide hydrolase (SEH) (Epoxide hydratase) (Cytosolic epoxide hydrolase) (cEH) Length = 286 Score = 41.6 bits (96), Expect = 0.006 Identities = 32/115 (27%), Positives = 51/115 (44%) Frame = +3 Query: 366 HRFTIIKGLQIHYIHVXXXXXXXXXXXXXXXXHGWPGSFFEFYKLIPLLSTQTEKDQYAF 545 H +I G ++HY+ HGWP S++E+ +IP L+ Q F Sbjct: 7 HHQAMINGYRMHYVTAGSGYPLVLL-------HGWPQSWYEWRNVIPALAEQ-------F 52 Query: 546 EIVCPSLVGYGWSDGATIPGFDLFAMSDVMFTLMERLNITSKFYIQGGDFGFIIA 710 ++ P L G G S+ + GFD M+ + L+ L K + G D+G +A Sbjct: 53 TVIAPDLRGLGDSE-KPMTGFDKRTMATDVRELVSHLGY-DKVGVIGHDWGGSVA 105
>sp|P53750|YN93_YEAST Hypothetical 32.8 kDa protein in BIO3-HXT17 intergenic region Length = 290 Score = 41.2 bits (95), Expect = 0.008 Identities = 28/97 (28%), Positives = 50/97 (51%) Frame = +3 Query: 462 HGWPGSFFEFYKLIPLLSTQTEKDQYAFEIVCPSLVGYGWSDGATIPGFDLFAMSDVMFT 641 HG+P S F LIPLL+ Q F I+ P L G+G+++ F ++ + + Sbjct: 36 HGFPTSSNMFRNLIPLLAGQ-------FHIIAPDLPGFGFTETPENYKFSFDSLCESIGY 88 Query: 642 LMERLNITSKFYIQGGDFGFIIARSMSVINPKKVAGI 752 L++ L+I KF + D+G + +++ P ++ GI Sbjct: 89 LLDTLSI-EKFAMYIFDYGSPVGFRLALKFPSRITGI 124
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 184,369,600
Number of Sequences: 369166
Number of extensions: 3948806
Number of successful extensions: 9806
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 9218
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 9760
length of database: 68,354,980
effective HSP length: 115
effective length of database: 47,110,455
effective search space used: 19221065640
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail