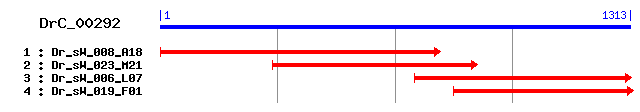
DrC_00292
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00292 (1313 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q94546|ATU_DROME Another transcription unit protein 136 2e-31 sp|P38439|LEO1_YEAST Protein LEO1 63 2e-09 sp|P56716|RP1_MOUSE Oxygen-regulated protein 1 (Retinitis p... 34 1.1 sp|Q9QX47|SON_MOUSE SON protein 33 1.4 sp|P18583|SON_HUMAN SON protein (SON3) (Negative regulatory... 33 1.8 sp|Q8DTQ5|HISZ_STRMU ATP phosphoribosyltransferase regulato... 33 2.4 sp|Q5SXM2|SNPC4_HUMAN snRNA-activating protein complex subu... 32 3.1 sp|Q03421|AROA_HAEIN 3-phosphoshikimate 1-carboxyvinyltrans... 31 6.9 sp|Q92WB5|UXUA_RHIME Mannonate dehydratase (D-mannonate hyd... 31 9.1 sp|Q8VDS8|STX18_MOUSE Syntaxin-18 31 9.1
>sp|Q94546|ATU_DROME Another transcription unit protein Length = 725 Score = 136 bits (342), Expect = 2e-31 Identities = 66/108 (61%), Positives = 79/108 (73%) Frame = +2 Query: 380 IEVDFPVIKADLGEEMHLVKLPNFLSVETRPFDPMYYEDEMDESEVQDEEGKTRLKLKVE 559 I+V+ P I ADLG+E H +KLPNFLSV T PFDP YEDE+DE E DEEG+ R+KLKV Sbjct: 380 IDVEIPRISADLGKEQHFIKLPNFLSVVTHPFDPETYEDEIDEEETMDEEGRQRIKLKVS 439 Query: 560 NTIRWRIAKDSEGQPILDENDQPKRESNARMIRWSDGSLSLHLGREIF 703 NTIRWR +++G + RESNAR +RWSDGS+SLHLG EIF Sbjct: 440 NTIRWREYMNNKGDMV--------RESNARFVRWSDGSMSLHLGNEIF 479
Score = 104 bits (260), Expect = 5e-22
Identities = 54/149 (36%), Positives = 87/149 (58%), Gaps = 5/149 (3%)
Frame = +3
Query: 690 GARYFDVHTTDISADHNHLFVREGSGLQAQAIFTSKLGFRPHSTDSFTHRKLTLSLADKT 869
G FD + + DHNHLFVR+G+GLQ Q++F +KL FRPHST+SFTH+K+T+SLAD++
Sbjct: 475 GNEIFDAYRQPLLGDHNHLFVRQGTGLQGQSVFRTKLTFRPHSTESFTHKKMTMSLADRS 534
Query: 870 HKAAKVKLLPFAGENPDVRQQEMIKREEERMKASLXXXXXXXXXXXXXXXXEL-----TA 1034
K + +K+L G++P + ++ EE +++ ++ E T+
Sbjct: 535 SKTSGIKILTQVGKDPTTDRPTQLREEEAKLRQAMRNQHKSLPKKKKPGAGEPLIGGGTS 594
Query: 1035 AFLEQQKDDENNPHAISLSDIKKKARPSS 1121
++ + D+ N AISLS IK + + S
Sbjct: 595 SYQHDEGSDDEN--AISLSAIKNRYKKGS 621
>sp|P38439|LEO1_YEAST Protein LEO1 Length = 464 Score = 62.8 bits (151), Expect = 2e-09 Identities = 32/94 (34%), Positives = 55/94 (58%), Gaps = 2/94 (2%) Frame = +2 Query: 422 EMHLVKLPNFLSVETRPFDPMYYEDEMDE--SEVQDEEGKTRLKLKVENTIRWRIAKDSE 595 E+ ++PNFL+++ PFDP +E +++E S E + +L ENT+RWR ++D + Sbjct: 167 EIFYARIPNFLTIDPIPFDPPSFEAKVNERASNSASREDQLDDRLIDENTVRWRYSRDKD 226 Query: 596 GQPILDENDQPKRESNARMIRWSDGSLSLHLGRE 697 +ESN ++++WSDG+ SL +G E Sbjct: 227 QHVF--------KESNTQIVQWSDGTYSLKVGEE 252
>sp|P56716|RP1_MOUSE Oxygen-regulated protein 1 (Retinitis pigmentosa RP1 protein homolog) Length = 2095 Score = 33.9 bits (76), Expect = 1.1 Identities = 21/75 (28%), Positives = 40/75 (53%), Gaps = 9/75 (12%) Frame = +2 Query: 446 NFLSVETRPFDPMYY---EDEMDESEVQDEEG------KTRLKLKVENTIRWRIAKDSEG 598 N+L++ET + ED++++S V +++G K R K+K E T++W + G Sbjct: 299 NYLALETHESQSLSTYPSEDDVEKSIVFNQDGTMTVVMKVRFKIKEEETVKWTTTVNRAG 358 Query: 599 QPILDENDQPKRESN 643 L ND+ ++S+ Sbjct: 359 ---LSNNDEKNKKSS 370
>sp|Q9QX47|SON_MOUSE SON protein Length = 2404 Score = 33.5 bits (75), Expect = 1.4 Identities = 20/58 (34%), Positives = 31/58 (53%), Gaps = 2/58 (3%) Frame = +1 Query: 1084 RCRISRRRRVPAPQSKRKEPSICPRRMRRKRSP--*AISQIPLRKRKERKLELSQTTK 1251 R R RRR +R+ SI P R+RR R+P S+ P+R+++ R E ++ K Sbjct: 1969 RSRTPSRRRRSRSAVRRRSFSISPVRLRRSRTPLRRRFSRSPIRRKRSRSSERGRSPK 2026
>sp|P18583|SON_HUMAN SON protein (SON3) (Negative regulatory element-binding protein) (NRE-binding protein) (DBP-5) (Bax antagonist selected in saccharomyces 1) (BASS1) Length = 2426 Score = 33.1 bits (74), Expect = 1.8 Identities = 20/58 (34%), Positives = 31/58 (53%), Gaps = 2/58 (3%) Frame = +1 Query: 1084 RCRISRRRRVPAPQSKRKEPSICPRRMRRKRSP--*AISQIPLRKRKERKLELSQTTK 1251 R R RRR +R+ SI P R+RR R+P S+ P+R+++ R E ++ K Sbjct: 1991 RSRTPSRRRRSRSVVRRRSFSISPVRLRRSRTPLRRRFSRSPIRRKRSRSSERGRSPK 2048
>sp|Q8DTQ5|HISZ_STRMU ATP phosphoribosyltransferase regulatory subunit Length = 320 Score = 32.7 bits (73), Expect = 2.4 Identities = 13/35 (37%), Positives = 24/35 (68%) Frame = +3 Query: 699 YFDVHTTDISADHNHLFVREGSGLQAQAIFTSKLG 803 +F+V D+S++H HLF ++G+ L + TS++G Sbjct: 47 HFEVFADDVSSEHYHLFDKKGNLLSLRPDITSQIG 81
>sp|Q5SXM2|SNPC4_HUMAN snRNA-activating protein complex subunit 4 (SNAPc subunit 4) (snRNA-activating protein complex 190 kDa subunit) (SNAPc 190 kDa subunit) (Proximal sequence element-binding transcription factor alpha subunit) (PSE-binding factor alpha subunit) (PTF alpha subunit) Length = 1469 Score = 32.3 bits (72), Expect = 3.1 Identities = 19/68 (27%), Positives = 30/68 (44%), Gaps = 1/68 (1%) Frame = +2 Query: 923 PPAGDDQARGGANEGLLASRESAQTDAGAREPPRTHRSVPRAAEGR*EQSSRYFAVGYQE 1102 PP G ++GG+ E + + + + P R H VPR+A+ +R Q Sbjct: 600 PPKGSSASQGGSKEASTTAAAPGEETSPVQVPARAHGPVPRSAQASHSADTRPAGAEKQA 659 Query: 1103 -EGASQLL 1123 EG +LL Sbjct: 660 LEGGRRLL 667
>sp|Q03421|AROA_HAEIN 3-phosphoshikimate 1-carboxyvinyltransferase (5-enolpyruvylshikimate-3-phosphate synthase) (EPSP synthase) (EPSPS) Length = 432 Score = 31.2 bits (69), Expect = 6.9 Identities = 17/36 (47%), Positives = 23/36 (63%) Frame = -1 Query: 863 VGERERQLAVRERVGRVGAEPELAGEDRLRLQPAPL 756 V E +R A+ + +VGAE E GED +R+QP PL Sbjct: 342 VKETDRLTAMATELRKVGAEVE-GGEDFIRIQPLPL 376
>sp|Q92WB5|UXUA_RHIME Mannonate dehydratase (D-mannonate hydrolase) Length = 402 Score = 30.8 bits (68), Expect = 9.1 Identities = 29/94 (30%), Positives = 40/94 (42%), Gaps = 10/94 (10%) Frame = +2 Query: 365 GDDGAIEV-DFPVIKADLGEEMHLVKLPNFLSVETRPFDPMYYEDEMDE---------SE 514 G GA+ D P + +H V L N L E R +YEDE E +E Sbjct: 273 GSLGALAANDLPAMTRQFASRIHFVHLRNVLREEVRT-PCSFYEDEHLEGDTDMVAVIAE 331 Query: 515 VQDEEGKTRLKLKVENTIRWRIAKDSEGQPILDE 616 + EE + R +V++ I R GQ ILD+ Sbjct: 332 LLQEEQRRRAAGRVDHQIPMR---PDHGQEILDD 362
>sp|Q8VDS8|STX18_MOUSE Syntaxin-18 Length = 308 Score = 30.8 bits (68), Expect = 9.1 Identities = 19/71 (26%), Positives = 31/71 (43%), Gaps = 1/71 (1%) Frame = +2 Query: 521 DEEGKTRLKLKVENTIRWRIAKDSEGQPILDE-NDQPKRESNARMIRWSDGSLSLHLGRE 697 + E T+ K +++DSEG+P +E ++P ES + W DG L E Sbjct: 144 EPEPHTKRKDSTSEKAPQNVSQDSEGKPAAEELPEKPLAESQPELGTWGDGKGEDELSPE 203 Query: 698 IFRRAHDRHQR 730 + +QR Sbjct: 204 EIQMFEQENQR 214
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.314 0.131 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 116,919,859
Number of Sequences: 369166
Number of extensions: 2134171
Number of successful extensions: 5170
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 4915
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5162
length of database: 68,354,980
effective HSP length: 114
effective length of database: 47,295,190
effective search space used: 15276346370
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
Cluster detail