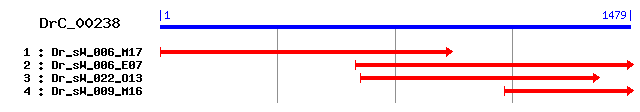
DrC_00238
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00238 (1478 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q9P289|MST4_HUMAN Serine/threonine-protein kinase MST4 (... 106 2e-22 sp|Q99JT2|MST4_MOUSE Serine/threonine-protein kinase MST4 (... 106 2e-22 sp|P23561|STE11_YEAST Serine/threonine-protein kinase STE11 105 4e-22 sp|P32490|MKK1_YEAST MAP kinase kinase MKK1/SSP32 104 8e-22 sp|Q9Y6E0|STK24_HUMAN Serine/threonine-protein kinase 24 (S... 102 3e-21 sp|P41892|CDC7_SCHPO Cell division control protein 7 102 4e-21 sp|Q9Y2U5|M3K2_HUMAN Mitogen-activated protein kinase kinas... 102 4e-21 sp|Q99KH8|STK24_MOUSE Serine/threonine-protein kinase 24 101 5e-21 sp|O00506|STK25_HUMAN Serine/threonine-protein kinase 25 (S... 100 8e-21 sp|O88643|PAK1_MOUSE Serine/threonine-protein kinase PAK 1 ... 100 8e-21
>sp|Q9P289|MST4_HUMAN Serine/threonine-protein kinase MST4 (STE20-like kinase MST4) (MST-4) (Mammalian STE20-like protein kinase 4) (Serine/threonine-protein kinase MASK) (Mst3 and SOK1-related kinase) Length = 416 Score = 106 bits (265), Expect = 2e-22 Identities = 78/256 (30%), Positives = 119/256 (46%), Gaps = 1/256 (0%) Frame = +2 Query: 626 EPFDTGMYSEIYKARDQVKNQSIVLKRY-ISDDGDLIEKYRKLINGLIDFQRKGGQERFV 802 E G + E++K D Q + +K + + D IE ++ I L Sbjct: 28 ERIGKGSFGEVFKGIDNRTQQVVAIKIIDLEEAEDEIEDIQQEITVLSQCD----SSYVT 83 Query: 803 KLYSFYPDPCSFAIGMEWMEGGSMDNYVKNFWPFTDKQLIRWTHQILGGLDFLHDNNVIF 982 K Y Y I ME++ GGS + ++ PF + Q+ +IL GLD+LH I Sbjct: 84 KYYGSYLKGSKLWIIMEYLGGGSALDLLRA-GPFDEFQIATMLKEILKGLDYLHSEKKIH 142 Query: 983 RDLKPSNVLLDENNNTKLCAFSLSIYLMSNHEMELKRSSSQSKVGVYSYMAPEVLNSEPY 1162 RD+K +NVLL E + KL F ++ L + ++KR++ VG +MAPEV+ Y Sbjct: 143 RDIKAANVLLSEQGDVKLADFGVAGQLT---DTQIKRNTF---VGTPFWMAPEVIQQSAY 196 Query: 1163 DNAADVWSFAAVLVNLITGKNPCPDKDPIDAAFHVARKLEMEYTLPEDTNQVLKDIITNC 1342 D+ AD+WS + L G+ P D P+ F + + TL D + K+ I C Sbjct: 197 DSKADIWSLGITAIELAKGEPPNSDMHPMRVLFLIPK--NNPPTLVGDFTKSFKEFIDAC 254 Query: 1343 AQGDPKKRPKIKELIE 1390 DP RP KEL++ Sbjct: 255 LNKDPSFRPTAKELLK 270
>sp|Q99JT2|MST4_MOUSE Serine/threonine-protein kinase MST4 (STE20-like kinase MST4) (MST-4) (Mammalian STE20-like protein kinase 4) Length = 416 Score = 106 bits (264), Expect = 2e-22 Identities = 78/256 (30%), Positives = 119/256 (46%), Gaps = 1/256 (0%) Frame = +2 Query: 626 EPFDTGMYSEIYKARDQVKNQSIVLKRY-ISDDGDLIEKYRKLINGLIDFQRKGGQERFV 802 E G + E++K D Q + +K + + D IE ++ I L Sbjct: 28 ERIGKGSFGEVFKGIDNRTQQVVAIKIIDLEEAEDEIEDIQQEITVLSQCD----SSYVT 83 Query: 803 KLYSFYPDPCSFAIGMEWMEGGSMDNYVKNFWPFTDKQLIRWTHQILGGLDFLHDNNVIF 982 K Y Y I ME++ GGS + ++ PF + Q+ +IL GLD+LH I Sbjct: 84 KYYGSYLKGSKLWIIMEYLGGGSALDLLRA-GPFDEFQIATMLKEILKGLDYLHSEKKIH 142 Query: 983 RDLKPSNVLLDENNNTKLCAFSLSIYLMSNHEMELKRSSSQSKVGVYSYMAPEVLNSEPY 1162 RD+K +NVLL E + KL F ++ L + ++KR++ VG +MAPEV+ Y Sbjct: 143 RDIKAANVLLSEQGDVKLADFGVAGQLT---DTQIKRNTF---VGTPFWMAPEVIQQSAY 196 Query: 1163 DNAADVWSFAAVLVNLITGKNPCPDKDPIDAAFHVARKLEMEYTLPEDTNQVLKDIITNC 1342 D+ AD+WS + L G+ P D P+ F + + TL D + K+ I C Sbjct: 197 DSKADIWSLGITAIELAKGEPPNSDMHPMRVLFLIPK--NNPPTLIGDFTKSFKEFIDAC 254 Query: 1343 AQGDPKKRPKIKELIE 1390 DP RP KEL++ Sbjct: 255 LNKDPSFRPTAKELLK 270
>sp|P23561|STE11_YEAST Serine/threonine-protein kinase STE11 Length = 738 Score = 105 bits (261), Expect = 4e-22 Identities = 63/220 (28%), Positives = 113/220 (51%), Gaps = 3/220 (1%) Frame = +2 Query: 740 YRKLINGL---IDFQRKGGQERFVKLYSFYPDPCSFAIGMEWMEGGSMDNYVKNFWPFTD 910 +RK+++ L ++ ++ E V Y + + I +E++ GGS+ + + N+ PF + Sbjct: 515 HRKMVDALQHEMNLLKELHHENIVTYYGASQEGGNLNIFLEYVPGGSVSSMLNNYGPFEE 574 Query: 911 KQLIRWTHQILGGLDFLHDNNVIFRDLKPSNVLLDENNNTKLCAFSLSIYLMSNHEMELK 1090 + +T QIL G+ +LH N+I RD+K +N+L+D K+ F +S L ++ + K Sbjct: 575 SLITNFTRQILIGVAYLHKKNIIHRDIKGANILIDIKGCVKITDFGISKKLSPLNKKQNK 634 Query: 1091 RSSSQSKVGVYSYMAPEVLNSEPYDNAADVWSFAAVLVNLITGKNPCPDKDPIDAAFHVA 1270 R+S Q V +M+PEV+ AD+WS V++ + TGK+P PD + A F + Sbjct: 635 RASLQGSV---FWMSPEVVKQTATTAKADIWSTGCVVIEMFTGKHPFPDFSQMQAIFKIG 691 Query: 1271 RKLEMEYTLPEDTNQVLKDIITNCAQGDPKKRPKIKELIE 1390 E +P K+ + + D + RP EL++ Sbjct: 692 TNTTPE--IPSWATSEGKNFLRKAFELDYQYRPSALELLQ 729
>sp|P32490|MKK1_YEAST MAP kinase kinase MKK1/SSP32 Length = 508 Score = 104 bits (259), Expect = 8e-22 Identities = 73/253 (28%), Positives = 114/253 (45%), Gaps = 15/253 (5%) Frame = +2 Query: 674 QVKNQSIVLKRYISDDGDLIEKYRKLINGLIDFQRKGGQERFVKLYSFYPDP--CSFAIG 847 ++KN S + + + + +Y+K I + F R E V+ Y + D S I Sbjct: 239 KLKNGSKIFALKVINTLNTDPEYQKQIFRELQFNRSFQSEYIVRYYGMFTDDENSSIYIA 298 Query: 848 MEWMEGGSMDNYVKNFWP----FTDKQLIRWTHQILGGLDFLHDNNVIFRDLKPSNVLLD 1015 ME+M G S+D KN ++K L + +L GL +LH+ VI RD+KP N+LL+ Sbjct: 299 MEYMGGRSLDAIYKNLLERGGRISEKVLGKIAEAVLRGLSYLHEKKVIHRDIKPQNILLN 358 Query: 1016 ENNNTKLCAFSLSIYLMSNHEMELKRSSSQSKVGVYSYMAPEVLNSEPYDNAADVWSFAA 1195 EN KLC F +S E S + + G YMAPE + +PY +DVWS Sbjct: 359 ENGQVKLCDFGVS--------GEAVNSLATTFTGTSFYMAPERIQGQPYSVTSDVWSLGL 410 Query: 1196 VLVNLITGKNPCPDKD------PIDAAFHVARKLEMEYTLPEDT---NQVLKDIITNCAQ 1348 ++ + GK PC + P + + PE + K I C + Sbjct: 411 TILEVANGKFPCSSEKMAANIAPFELLMWILTFTPELKDEPESNIIWSPSFKSFIDYCLK 470 Query: 1349 GDPKKRPKIKELI 1387 D ++RP +++I Sbjct: 471 KDSRERPSPRQMI 483
>sp|Q9Y6E0|STK24_HUMAN Serine/threonine-protein kinase 24 (STE20-like kinase MST3) (MST-3) (Mammalian STE20-like protein kinase 3) Length = 443 Score = 102 bits (254), Expect = 3e-21 Identities = 79/273 (28%), Positives = 126/273 (46%), Gaps = 12/273 (4%) Frame = +2 Query: 608 DVKIDPEPFDT-------GMYSEIYKARDQVKNQSIVLKRY-ISDDGDLIEKYRKLINGL 763 ++K DPE T G + E++K D + + +K + + D IE ++ I L Sbjct: 27 NLKADPEELFTKLEKIGKGSFGEVFKGIDNRTQKVVAIKIIDLEEAEDEIEDIQQEITVL 86 Query: 764 IDFQRKGGQERFVKLYSFYPDPCSFAIGMEWMEGGSMDNYVKNFWPFTDKQLIRWTHQIL 943 K Y Y I ME++ GGS + ++ P + Q+ +IL Sbjct: 87 SQCD----SPYVTKYYGSYLKDTKLWIIMEYLGGGSALDLLEP-GPLDETQIATILREIL 141 Query: 944 GGLDFLHDNNVIFRDLKPSNVLLDENNNTKLCAFSLSIYLMSNHEMELKRSSSQSKVGVY 1123 GLD+LH I RD+K +NVLL E+ KL F ++ L + ++KR++ VG Sbjct: 142 KGLDYLHSEKKIHRDIKAANVLLSEHGEVKLADFGVAGQLT---DTQIKRNTF---VGTP 195 Query: 1124 SYMAPEVLNSEPYDNAADVWSFAAVLVNLITGKNPCPDKDPIDAAFHVARK----LEMEY 1291 +MAPEV+ YD+ AD+WS + L G+ P + P+ F + + LE Y Sbjct: 196 FWMAPEVIKQSAYDSKADIWSLGITAIELARGEPPHSELHPMKVLFLIPKNNPPTLEGNY 255 Query: 1292 TLPEDTNQVLKDIITNCAQGDPKKRPKIKELIE 1390 + P LK+ + C +P RP KEL++ Sbjct: 256 SKP------LKEFVEACLNKEPSFRPTAKELLK 282
>sp|P41892|CDC7_SCHPO Cell division control protein 7 Length = 1062 Score = 102 bits (253), Expect = 4e-21 Identities = 65/250 (26%), Positives = 115/250 (46%) Frame = +2 Query: 641 GMYSEIYKARDQVKNQSIVLKRYISDDGDLIEKYRKLINGLIDFQRKGGQERFVKLYSFY 820 G + +Y+ + +++ +K+ +++ +I ID + VK Y Sbjct: 18 GAFGAVYRGLNIKNGETVAVKKVKLSK--MLKSDLSVIKMEIDLLKNLDHPNIVKYRGSY 75 Query: 821 PDPCSFAIGMEWMEGGSMDNYVKNFWPFTDKQLIRWTHQILGGLDFLHDNNVIFRDLKPS 1000 S I +E+ E GS+ + KNF + + +T Q+L GL +LH+ VI RD+K + Sbjct: 76 QTNDSLCIILEYCENGSLRSICKNFGKIPENLVALYTFQVLQGLLYLHNQGVIHRDIKGA 135 Query: 1001 NVLLDENNNTKLCAFSLSIYLMSNHEMELKRSSSQSKVGVYSYMAPEVLNSEPYDNAADV 1180 N+L ++ KL F ++ ++ S VG +MAPEV+ A+D+ Sbjct: 136 NILTTKDGTIKLADFGVA--------TKINALEDHSVVGSPYWMAPEVIELVGATTASDI 187 Query: 1181 WSFAAVLVNLITGKNPCPDKDPIDAAFHVARKLEMEYTLPEDTNQVLKDIITNCAQGDPK 1360 WS ++ L+ G P D DP A F + + + LP + + K + C Q DP Sbjct: 188 WSVGCTVIELLDGNPPYYDLDPTSALFRMVK--DEHPPLPSNISSAAKSFLMQCFQKDPN 245 Query: 1361 KRPKIKELIE 1390 R K ++L++ Sbjct: 246 LRIKTRKLLK 255
>sp|Q9Y2U5|M3K2_HUMAN Mitogen-activated protein kinase kinase kinase 2 (MAPK/ERK kinase kinase 2) (MEK kinase 2) (MEKK 2) Length = 618 Score = 102 bits (253), Expect = 4e-21 Identities = 73/254 (28%), Positives = 125/254 (49%), Gaps = 5/254 (1%) Frame = +2 Query: 641 GMYSEIYKARDQVKNQSIVLKRYISDDGDLIEKYRKLINGL---IDFQRKGGQERFVKLY 811 G + +Y D + + +K+ + D D E ++ +N L I + ER V+ Y Sbjct: 364 GAFGRVYLCYDVDTGRELAVKQ-VQFDPDSPETSKE-VNALECEIQLLKNFLHERIVQYY 421 Query: 812 SFYPDPC--SFAIGMEWMEGGSMDNYVKNFWPFTDKQLIRWTHQILGGLDFLHDNNVIFR 985 DP + +I ME+M GGS+ + +K + T+ ++T QIL G+ +LH N ++ R Sbjct: 422 GCLRDPQEKTLSIFMEYMPGGSIKDQLKAYGALTENGTRKYTRQILEGVHYLHSNMILHR 481 Query: 986 DLKPSNVLLDENNNTKLCAFSLSIYLMSNHEMELKRSSSQSKVGVYSYMAPEVLNSEPYD 1165 D+K +N+L D N KL F S L + + L + +S G +M+PEV++ + Y Sbjct: 482 DIKGANILRDSTGNVKLGDFGASKRLQT---ICLSGTGMKSVTGTPYWMSPEVISGQGYG 538 Query: 1166 NAADVWSFAAVLVNLITGKNPCPDKDPIDAAFHVARKLEMEYTLPEDTNQVLKDIITNCA 1345 AD+WS A +V ++T K P + + + A F +A + LP + +D + Sbjct: 539 RKADIWSVACTVVEMLTEKPPWAEFEAMAAIFKIATQ-PTNPKLPPHVSDYTRDFLKRIF 597 Query: 1346 QGDPKKRPKIKELI 1387 + K RP EL+ Sbjct: 598 V-EAKLRPSADELL 610
>sp|Q99KH8|STK24_MOUSE Serine/threonine-protein kinase 24 Length = 431 Score = 101 bits (252), Expect = 5e-21 Identities = 79/273 (28%), Positives = 126/273 (46%), Gaps = 12/273 (4%) Frame = +2 Query: 608 DVKIDPEPFDT-------GMYSEIYKARDQVKNQSIVLKRY-ISDDGDLIEKYRKLINGL 763 ++K DPE T G + E++K D + + +K + + D IE ++ I L Sbjct: 15 NLKADPEELFTKLEKIGKGSFGEVFKGIDNRTQKVVAIKIIDLEEAEDEIEDIQQEITVL 74 Query: 764 IDFQRKGGQERFVKLYSFYPDPCSFAIGMEWMEGGSMDNYVKNFWPFTDKQLIRWTHQIL 943 K Y Y I ME++ GGS + ++ P + Q+ +IL Sbjct: 75 SQCD----SPYVTKYYGSYLKDTKLWIIMEYLGGGSALDLLEP-GPLDEIQIATILREIL 129 Query: 944 GGLDFLHDNNVIFRDLKPSNVLLDENNNTKLCAFSLSIYLMSNHEMELKRSSSQSKVGVY 1123 GLD+LH I RD+K +NVLL E+ KL F ++ L + ++KR++ VG Sbjct: 130 KGLDYLHSEKKIHRDIKAANVLLSEHGEVKLADFGVAGQLT---DTQIKRNTF---VGTP 183 Query: 1124 SYMAPEVLNSEPYDNAADVWSFAAVLVNLITGKNPCPDKDPIDAAFHVARK----LEMEY 1291 +MAPEV+ YD+ AD+WS + L G+ P + P+ F + + LE Y Sbjct: 184 FWMAPEVIKQSAYDSKADIWSLGITAIELAKGEPPHSELHPMKVLFLIPKNNPPTLEGNY 243 Query: 1292 TLPEDTNQVLKDIITNCAQGDPKKRPKIKELIE 1390 + P LK+ + C +P RP KEL++ Sbjct: 244 SKP------LKEFVEACLNKEPSFRPTAKELLK 270
>sp|O00506|STK25_HUMAN Serine/threonine-protein kinase 25 (Sterile 20/oxidant stress-response kinase 1) (Ste20/oxidant stress response kinase-1) (SOK-1) (Ste20-like kinase) Length = 426 Score = 100 bits (250), Expect = 8e-21 Identities = 76/267 (28%), Positives = 122/267 (45%), Gaps = 8/267 (2%) Frame = +2 Query: 614 KIDPEPFDT-------GMYSEIYKARDQVKNQSIVLKRY-ISDDGDLIEKYRKLINGLID 769 ++DPE T G + E+YK D + + +K + + D IE ++ I L Sbjct: 13 RVDPEELFTKLDRIGKGSFGEVYKGIDNHTKEVVAIKIIDLEEAEDEIEDIQQEITVLSQ 72 Query: 770 FQRKGGQERFVKLYSFYPDPCSFAIGMEWMEGGSMDNYVKNFWPFTDKQLIRWTHQILGG 949 + + Y I ME++ GGS + +K P + + +IL G Sbjct: 73 CD----SPYITRYFGSYLKSTKLWIIMEYLGGGSALDLLKP-GPLEETYIATILREILKG 127 Query: 950 LDFLHDNNVIFRDLKPSNVLLDENNNTKLCAFSLSIYLMSNHEMELKRSSSQSKVGVYSY 1129 LD+LH I RD+K +NVLL E + KL F ++ L + ++KR++ VG + Sbjct: 128 LDYLHSERKIHRDIKAANVLLSEQGDVKLADFGVAGQLT---DTQIKRNTF---VGTPFW 181 Query: 1130 MAPEVLNSEPYDNAADVWSFAAVLVNLITGKNPCPDKDPIDAAFHVARKLEMEYTLPEDT 1309 MAPEV+ YD AD+WS + L G+ P D P+ F + + TL Sbjct: 182 MAPEVIKQSAYDFKADIWSLGITAIELAKGEPPNSDLHPMRVLFLIPK--NSPPTLEGQH 239 Query: 1310 NQVLKDIITNCAQGDPKKRPKIKELIE 1390 ++ K+ + C DP+ RP KEL++ Sbjct: 240 SKPFKEFVEACLNKDPRFRPTAKELLK 266
>sp|O88643|PAK1_MOUSE Serine/threonine-protein kinase PAK 1 (p21-activated kinase 1) (PAK-1) (P65-PAK) (Alpha-PAK) (CDC42/RAC effector kinase PAK-A) Length = 545 Score = 100 bits (250), Expect = 8e-21 Identities = 76/298 (25%), Positives = 131/298 (43%), Gaps = 3/298 (1%) Frame = +2 Query: 524 NEKKHQRNPFIYPYINLEDSFIKLMKIPDVKIDPEPFDT---GMYSEIYKARDQVKNQSI 694 N +K ++ P + LE ++ + D K PF+ G +Y A D Q + Sbjct: 238 NTEKQKKKPKMSDEEILE-KLRSIVSVGDPKKKYTPFEKIGQGASGTVYTAMDVATGQEV 296 Query: 695 VLKRYISDDGDLIEKYRKLINGLIDFQRKGGQERFVKLYSFYPDPCSFAIGMEWMEGGSM 874 +K+ + ++LI I R+ V Y + ME++ GGS+ Sbjct: 297 AIKQMNLQQ----QPKKELIINEILVMRENKNPNIVNYLDSYLVGDELWVVMEYLAGGSL 352 Query: 875 DNYVKNFWPFTDKQLIRWTHQILGGLDFLHDNNVIFRDLKPSNVLLDENNNTKLCAFSLS 1054 + V + Q+ + L L+FLH N VI RD+K N+LL + + KL F Sbjct: 353 TDVVTETC-MDEGQIAAVCRECLQALEFLHSNQVIHRDIKSDNILLGMDGSVKLTDFGFC 411 Query: 1055 IYLMSNHEMELKRSSSQSKVGVYSYMAPEVLNSEPYDNAADVWSFAAVLVNLITGKNPCP 1234 + ++S + VG +MAPEV+ + Y D+WS + + +I G+ P Sbjct: 412 AQITP------EQSKRSTMVGTPYWMAPEVVTRKAYGPKVDIWSLGIMAIEMIEGEPPYL 465 Query: 1235 DKDPIDAAFHVARKLEMEYTLPEDTNQVLKDIITNCAQGDPKKRPKIKELIEIDLQKI 1408 +++P+ A + +A E PE + + +D + C + D +KR KEL++ KI Sbjct: 466 NENPLRALYLIATNGTPELQNPEKLSAIFRDFLQCCLEMDVEKRGSAKELLQHQFLKI 523
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 164,282,747
Number of Sequences: 369166
Number of extensions: 3441047
Number of successful extensions: 12670
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 10211
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10985
length of database: 68,354,980
effective HSP length: 115
effective length of database: 47,110,455
effective search space used: 17760641535
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail