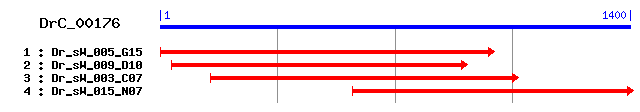
DrC_00176
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00176 (1400 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q26609|HXK_SCHMA Hexokinase 489 e-137 sp|P52789|HXK2_HUMAN Hexokinase, type II (HK II) (Muscle fo... 357 5e-98 sp|P05708|HXK1_RAT Hexokinase, type I (HK I) (Brain form he... 356 1e-97 sp|O08528|HXK2_MOUSE Hexokinase type II (HK II) 352 2e-96 sp|P17710|HXK1_MOUSE Hexokinase type I (HK I) (Hexokinase, ... 352 2e-96 sp|P27881|HXK2_RAT Hexokinase type II (HK II) 351 3e-96 sp|Q9NFT7|HXK2_DROME Hexokinase type 2 347 5e-95 sp|P35557|HXK4_HUMAN Hexokinase D (Hexokinase type IV) (HK ... 345 2e-94 sp|P27595|HXK1_BOVIN Hexokinase type I (HK I) (Brain form h... 345 2e-94 sp|P17712|HXK4_RAT Hexokinase D (Hexokinase type IV) (HK IV... 343 7e-94
>sp|Q26609|HXK_SCHMA Hexokinase Length = 451 Score = 489 bits (1258), Expect = e-137 Identities = 248/422 (58%), Positives = 311/422 (73%), Gaps = 1/422 (0%) Frame = +1 Query: 1 DLKFNDYENIMHLLEENMIRGLSAATHKDASVKMFPSYVTKLPTGKEEGQFLALDLGGTN 180 DL DYE I + E+M GL +T++ +S+KMFPSYVTK P G E G FLALDLGGTN Sbjct: 20 DLSVVDYEEICDRMGESMRLGLQKSTNEKSSIKMFPSYVTKTPNGTETGNFLALDLGGTN 79 Query: 181 YRVLLVTLR-QNEKPDIKETNYAIPKDKMTGKGEELFDYIAKTLSDFIKIHQIEKWPIKL 357 YRVL VTL + + P I+E Y IP +KM+G G ELF YIA+TL+DF++ + ++ L Sbjct: 80 YRVLSVTLEGKGKSPRIQERTYCIPAEKMSGSGTELFKYIAETLADFLENNGMKDKKFDL 139 Query: 358 GFTFSFPCSQINLSKALLVRWTKGFTASDCEGQDVAALLQNAINKQNLNVKCVAVVNDTV 537 GFTFSFPC Q L+ A LVRWTKGF+A EG +VA LLQ ++K+ LNVKCVAVVNDTV Sbjct: 140 GFTFSFPCVQKGLTHATLVRWTKGFSADGVEGHNVAELLQTELDKRELNVKCVAVVNDTV 199 Query: 538 GTLTSCALQDPKCGVGVIVGTGTNASYIEKIERVETFTEDKTGVEYVIINMEWGAFGDDG 717 GTL SCAL+DPKC VG+IVGTGTN +YIE +VE D V+IN EWGAFG+ G Sbjct: 200 GTLASCALEDPKCAVGLIVGTGTNVAYIEDSSKVELM--DGVKEPEVVINTEWGAFGEKG 257 Query: 718 CLDKYRTKYDKLLDEQSLHPNQQIFEKMCSGMYLGEIVRLVLLDLIDRKILFRGEKPKSL 897 LD +RT++DK +D SLHP +Q++EKM SGMYLGE+VR +++ L+++KILFRG+ P+ L Sbjct: 258 ELDCWRTQFDKSMDIDSLHPGKQLYEKMVSGMYLGELVRHIIVYLVEQKILFRGDLPERL 317 Query: 898 EASGSFHTKYLSEVERDPPHLLYCTSYMLTEDFNVPTFEEKDAAIVRAVCEKVAARAAFY 1077 + S T+YL++VERDP HLLY T YMLT+D +VP E D IVR CE V RAA+ Sbjct: 318 KVRNSLLTRYLTDVERDPAHLLYNTHYMLTDDLHVPVVEPIDNRIVRYACEMVVKRAAYL 377 Query: 1078 IGTGIACLLNRINKKEVTVGIDGSLFKFHCKFPERMTDIVHQLKNENIKFVLKLSEDGSG 1257 G GIAC+L RIN+ EVTVG+DGSL+KFH KF ERMTD+V +LK +N +F L+LSEDGSG Sbjct: 378 AGAGIACILRRINRSEVTVGVDGSLYKFHPKFCERMTDMVDKLKPKNTRFCLRLSEDGSG 437 Query: 1258 KG 1263 KG Sbjct: 438 KG 439
>sp|P52789|HXK2_HUMAN Hexokinase, type II (HK II) (Muscle form hexokinase) Length = 917 Score = 357 bits (916), Expect = 5e-98 Identities = 197/418 (47%), Positives = 276/418 (66%), Gaps = 10/418 (2%) Frame = +1 Query: 40 LEENMIRGLSAATHKDASVKMFPSYVTKLPTGKEEGQFLALDLGGTNYRVLLVTLRQNEK 219 ++ M RGLS TH A VKM P+YV P G E+G FLALDLGGTN+RVLLV +R + Sbjct: 491 MKVEMERGLSKETHASAPVKMLPTYVCATPDGTEKGDFLALDLGGTNFRVLLVRVRNGKW 550 Query: 220 PDIKETN--YAIPKDKMTGKGEELFDYIAKTLSDFIKIHQIEKWPIKLGFTFSFPCSQIN 393 ++ N YAIP++ M G G+ELFD+I + ++DF++ ++ + LGFTFSFPC Q + Sbjct: 551 GGVEMHNKIYAIPQEVMHGTGDELFDHIVQCIADFLEYMGMKGVSLPLGFTFSFPCQQNS 610 Query: 394 LSKALLVRWTKGFTASDCEGQDVAALLQNAINK-QNLNVKCVAVVNDTVGTLTSCALQDP 570 L +++L++WTKGF AS CEG+DV LL+ AI++ + ++ VAVVNDTVGT+ +C +DP Sbjct: 611 LDESILLKWTKGFKASGCEGEDVVTLLKEAIHRREEFDLDVVAVVNDTVGTMMTCGFEDP 670 Query: 571 KCGVGVIVGTGTNASYIEKIERVETFTEDKTGVEYVIINMEWGAFGDDGCLDKYRTKYDK 750 C VG+IVGTG+NA Y+E++ VE E + G + +NMEWGAFGD+GCLD +RT++D Sbjct: 671 HCEVGLIVGTGSNACYMEEMRNVE-LVEGEEG--RMCVNMEWGAFGDNGCLDDFRTEFDV 727 Query: 751 LLDEQSLHPNQQIFEKMCSGMYLGEIVRLVLLDLIDRKILFRGEKPKSLEASGSFHTKYL 930 +DE SL+P +Q FEKM SGMYLGEIVR +L+D R +LFRG + L+ G F TK+L Sbjct: 728 AVDELSLNPGKQRFEKMISGMYLGEIVRNILIDFTKRGLLFRGRISERLKTRGIFETKFL 787 Query: 931 SEVERDPPHLLYCTSYMLTEDFNVPTFEEKDAAIVRAVCEKVAARAAFYIGTGIACLLNR 1110 S++E D LL + + D+ IV+ VC VA RAA G G+A +++R Sbjct: 788 SQIESDCLALLQVRAILQHLGLESTC---DDSIIVKEVCTVVARRAAQLCGAGMAAVVDR 844 Query: 1111 INKK------EVTVGIDGSLFKFHCKFPERMTDIVHQLKNE-NIKFVLKLSEDGSGKG 1263 I + +VTVG+DG+L+K H F + M + V L + ++ F+ SEDGSGKG Sbjct: 845 IRENRGLDALKVTVGVDGTLYKLHPHFAKVMHETVKDLAPKCDVSFL--QSEDGSGKG 900
Score = 323 bits (828), Expect = 7e-88
Identities = 179/416 (43%), Positives = 267/416 (64%), Gaps = 10/416 (2%)
Frame = +1
Query: 46 ENMIRGLSAATHKDASVKMFPSYVTKLPTGKEEGQFLALDLGGTNYRVLLVTLRQN--EK 219
+ M +GL A TH A+VKM P++V P G E G+FLALDLGGTN+RVL V + N +K
Sbjct: 45 KEMEKGLGATTHPTAAVKMLPTFVRSTPDGTEHGEFLALDLGGTNFRVLWVKVTDNGLQK 104
Query: 220 PDIKETNYAIPKDKMTGKGEELFDYIAKTLSDFIKIHQIEKWPIKLGFTFSFPCSQINLS 399
+++ YAIP+D M G G +LFD+IA+ L++F+ QI+ + LGFTFSFPC Q L
Sbjct: 105 VEMENQIYAIPEDIMRGSGTQLFDHIAECLANFMDKLQIKDKKLPLGFTFSFPCHQTKLD 164
Query: 400 KALLVRWTKGFTASDCEGQDVAALLQNAINKQ-NLNVKCVAVVNDTVGTLTSCALQDPKC 576
++ LV WTKGF +S EG+DV AL++ AI ++ + ++ VAVVNDTVGT+ +C D C
Sbjct: 165 ESFLVSWTKGFKSSGVEGRDVVALIRKAIQRRGDFDIDIVAVVNDTVGTMMTCGYDDHNC 224
Query: 577 GVGVIVGTGTNASYIEKIERVETFTEDKTGVEYVIINMEWGAFGDDGCLDKYRTKYDKLL 756
+G+IVGTG+NA Y+E++ ++ D+ + INMEWGAFGDDG L+ RT++D+ +
Sbjct: 225 EIGLIVGTGSNACYMEEMRHIDMVEGDEGRM---CINMEWGAFGDDGSLNDIRTEFDQEI 281
Query: 757 DEQSLHPNQQIFEKMCSGMYLGEIVRLVLLDLIDRKILFRGEKPKSLEASGSFHTKYLSE 936
D SL+P +Q+FEKM SGMY+GE+VRL+L+ + ++LF G+ L +G F TK +S+
Sbjct: 282 DMGSLNPGKQLFEKMISGMYMGELVRLILVKMAKEELLFGGKLSPELLNTGRFETKDISD 341
Query: 937 VERDPPHLLYCTSYMLTEDFNVPTFEEKDAAIVRAVCEKVAARAAFYIGTGIACLLNRI- 1113
+E + + ++ + PT ++D +C+ V+ R+A +A +L RI
Sbjct: 342 IEGEKDGIRKAREVLMRLGLD-PT--QEDCVATHRICQIVSTRSASLCAATLAAVLQRIK 398
Query: 1114 -NKKE----VTVGIDGSLFKFHCKFPERMTDIVHQL-KNENIKFVLKLSEDGSGKG 1263
NK E T+G+DGS++K H F +R+ V +L +++F+ SEDGSGKG
Sbjct: 399 ENKGEERLRSTIGVDGSVYKKHPHFAKRLHKTVRRLVPGCDVRFL--RSEDGSGKG 452
>sp|P05708|HXK1_RAT Hexokinase, type I (HK I) (Brain form hexokinase) Length = 918 Score = 356 bits (913), Expect = 1e-97 Identities = 197/418 (47%), Positives = 269/418 (64%), Gaps = 10/418 (2%) Frame = +1 Query: 40 LEENMIRGLSAATHKDASVKMFPSYVTKLPTGKEEGQFLALDLGGTNYRVLLVTLRQNEK 219 L M GL T+ A+VKM PS+V +P G E G FLALDLGGTN+RVLLV +R +K Sbjct: 491 LRTEMEMGLRKETNSKATVKMLPSFVRSIPDGTEHGDFLALDLGGTNFRVLLVKIRSGKK 550 Query: 220 PDIKETN--YAIPKDKMTGKGEELFDYIAKTLSDFIKIHQIEKWPIKLGFTFSFPCSQIN 393 ++ N Y+IP + M G G+ELFD+I +SDF+ I+ + LGFTFSFPC Q N Sbjct: 551 RTVEMHNKIYSIPLEIMQGTGDELFDHIVSCISDFLDYMGIKGPRMPLGFTFSFPCHQTN 610 Query: 394 LSKALLVRWTKGFTASDCEGQDVAALLQNAINK-QNLNVKCVAVVNDTVGTLTSCALQDP 570 L +L+ WTKGF A+DCEG DVA+LL++A+ + + ++ VAVVNDTVGT+ +CA ++P Sbjct: 611 LDCGILISWTKGFKATDCEGHDVASLLRDAVKRREEFDLDVVAVVNDTVGTMMTCAYEEP 670 Query: 571 KCGVGVIVGTGTNASYIEKIERVETFTEDKTGVEYVIINMEWGAFGDDGCLDKYRTKYDK 750 C +G+IVGTGTNA Y+E+++ VE E G + INMEWGAFGD+GCLD RT +DK Sbjct: 671 TCEIGLIVGTGTNACYMEEMKNVE-MVEGNQG--QMCINMEWGAFGDNGCLDDIRTDFDK 727 Query: 751 LLDEQSLHPNQQIFEKMCSGMYLGEIVRLVLLDLIDRKILFRGEKPKSLEASGSFHTKYL 930 ++DE SL+ +Q FEKM SGMYLGEIVR +L+D + LFRG+ + L+ G F TK+L Sbjct: 728 VVDEYSLNSGKQRFEKMISGMYLGEIVRNILIDFTKKGFLFRGQISEPLKTRGIFETKFL 787 Query: 931 SEVERDPPHLLYCTSYMLTEDFNVPTFEEKDAAIVRAVCEKVAARAAFYIGTGIACLLNR 1110 S++E D LL + + N D+ +V+ VC V+ RAA G G+A ++ + Sbjct: 788 SQIESDRLALLQVRAILQQLGLNSTC---DDSILVKTVCGVVSKRAAQLCGAGMAAVVEK 844 Query: 1111 INKK------EVTVGIDGSLFKFHCKFPERMTDIVHQLKNE-NIKFVLKLSEDGSGKG 1263 I + VTVG+DG+L+K H F M V +L + + F+ LSEDGSGKG Sbjct: 845 IRENRGLDHLNVTVGVDGTLYKLHPHFSRIMHQTVKELSPKCTVSFL--LSEDGSGKG 900
Score = 300 bits (768), Expect = 7e-81
Identities = 166/423 (39%), Positives = 258/423 (60%), Gaps = 10/423 (2%)
Frame = +1
Query: 25 NIMHLLEENMIRGLSAATHKDASVKMFPSYVTKLPTGKEEGQFLALDLGGTNYRVLLVTL 204
+I+ ++ M GLS + ASVKM P++V +P G E+G F+ALDLGG+++R+L V +
Sbjct: 38 DILTRFKKEMKNGLSRDYNPTASVKMLPTFVRSIPDGSEKGDFIALDLGGSSFRILRVQV 97
Query: 205 R--QNEKPDIKETNYAIPKDKMTGKGEELFDYIAKTLSDFIKIHQIEKWPIKLGFTFSFP 378
+N+ ++ Y P++ + G G +LFD++A L DF++ +I+ + +GFTFSFP
Sbjct: 98 NHEKNQNVSMESEIYDTPENIVHGSGTQLFDHVADCLGDFMEKKKIKDKKLPVGFTFSFP 157
Query: 379 CSQINLSKALLVRWTKGFTASDCEGQDVAALLQNAINKQ-NLNVKCVAVVNDTVGTLTSC 555
C Q + +A+L+ WTK F AS EG DV LL AI K+ + + VAVVNDTVGT+ +C
Sbjct: 158 CRQSKIDEAVLITWTKRFKASGVEGADVVKLLNKAIKKRGDYDANIVAVVNDTVGTMMTC 217
Query: 556 ALQDPKCGVGVIVGTGTNASYIEKIERVETFTEDKTGVEYVIINMEWGAFGDDGCLDKYR 735
D +C VG+I+GTGTNA Y+E++ ++ D+ + IN EWGAFGDDG L+ R
Sbjct: 218 GYDDQQCEVGLIIGTGTNACYMEELRHIDLVEGDEGRM---CINTEWGAFGDDGSLEDIR 274
Query: 736 TKYDKLLDEQSLHPNQQIFEKMCSGMYLGEIVRLVLLDLIDRKILFRGEKPKSLEASGSF 915
T++D+ LD SL+P +Q+FEKM SGMY+GE+VRL+L+ + +LF G L G F
Sbjct: 275 TEFDRELDRGSLNPGKQLFEKMVSGMYMGELVRLILVKMAKEGLLFEGRITPELLTRGKF 334
Query: 916 HTKYLSEVERDPPHLLYCTSYMLTEDFNVPTFEEKDAAIVRAVCEKVAARAAFYIGTGIA 1095
+T +S +E+D + +LT P+ + D V+ +C V+ R+A + +
Sbjct: 335 NTSDVSAIEKDKEGIQNAKE-ILTRLGVEPS--DVDCVSVQHICTIVSFRSANLVAATLG 391
Query: 1096 CLLNRIN------KKEVTVGIDGSLFKFHCKFPERMTDIVHQL-KNENIKFVLKLSEDGS 1254
+LNR+ + TVG+DGSL+K H ++ R + +L + +++F+ LSE G+
Sbjct: 392 AILNRLRDNKGTPRLRTTVGVDGSLYKMHPQYSRRFHKTLRRLVPDSDVRFL--LSESGT 449
Query: 1255 GKG 1263
GKG
Sbjct: 450 GKG 452
>sp|O08528|HXK2_MOUSE Hexokinase type II (HK II) Length = 917 Score = 352 bits (902), Expect = 2e-96 Identities = 193/418 (46%), Positives = 274/418 (65%), Gaps = 10/418 (2%) Frame = +1 Query: 40 LEENMIRGLSAATHKDASVKMFPSYVTKLPTGKEEGQFLALDLGGTNYRVLLVTLRQNEK 219 ++ M +GLS TH+ A VKM P+YV P G E+G FLALDLGGTN+RVLLV +R ++ Sbjct: 491 MKVEMEQGLSKETHEAAPVKMLPTYVCATPDGTEKGDFLALDLGGTNFRVLLVRVRNGKR 550 Query: 220 PDIKETN--YAIPKDKMTGKGEELFDYIAKTLSDFIKIHQIEKWPIKLGFTFSFPCSQIN 393 ++ N Y+IP++ M G GEELFD+I + ++DF++ ++ + LGFTFSFPC Q + Sbjct: 551 RGVEMHNKIYSIPQEVMHGTGEELFDHIVQCIADFLEYMGMKGVSLPLGFTFSFPCQQNS 610 Query: 394 LSKALLVRWTKGFTASDCEGQDVAALLQNAINK-QNLNVKCVAVVNDTVGTLTSCALQDP 570 L +++L++WTKGF AS CEG+DV LL+ AI + + ++ VAVVNDTVGT+ +C +DP Sbjct: 611 LDQSILLKWTKGFKASGCEGEDVVTLLKEAIRRREEFDLDVVAVVNDTVGTMMTCGYEDP 670 Query: 571 KCGVGVIVGTGTNASYIEKIERVETFTEDKTGVEYVIINMEWGAFGDDGCLDKYRTKYDK 750 C VG+IVGTG+NA Y+E++ VE ++ + +NMEWGAFGD+GCLD RT +D Sbjct: 671 HCEVGLIVGTGSNACYMEEMRNVELVDGEEGRM---CVNMEWGAFGDNGCLDDLRTVFDV 727 Query: 751 LLDEQSLHPNQQIFEKMCSGMYLGEIVRLVLLDLIDRKILFRGEKPKSLEASGSFHTKYL 930 +DE SL+P +Q FEKM SGMYLGEIVR +L+D R +LFRG + L+ G F TK+L Sbjct: 728 AVDELSLNPGKQRFEKMISGMYLGEIVRNILIDFTKRGLLFRGRISERLKTRGIFETKFL 787 Query: 931 SEVERDPPHLLYCTSYMLTEDFNVPTFEEKDAAIVRAVCEKVAARAAFYIGTGIACLLNR 1110 S++E D LL + + D+ IV+ VC VA RAA G G+A ++++ Sbjct: 788 SQIESDCLALLQVRAILRHLGLESTC---DDSIIVKEVCTVVARRAAQLCGAGMAAVVDK 844 Query: 1111 INKK------EVTVGIDGSLFKFHCKFPERMTDIVHQLKNE-NIKFVLKLSEDGSGKG 1263 I + +VTVG+DG+L+K H F + M + V L + ++ F+ SEDGSGKG Sbjct: 845 IRENRGLDNLKVTVGVDGTLYKLHPHFAKVMHETVRDLAPKCDVSFL--ESEDGSGKG 900
Score = 325 bits (834), Expect = 1e-88
Identities = 180/422 (42%), Positives = 270/422 (63%), Gaps = 10/422 (2%)
Frame = +1
Query: 28 IMHLLEENMIRGLSAATHKDASVKMFPSYVTKLPTGKEEGQFLALDLGGTNYRVLLVTLR 207
I + M +GL A TH A+VKM P++V P G E G+FLALDLGGTN+RVL V +
Sbjct: 39 ISRRFRKEMEKGLGATTHPTAAVKMLPTFVRSTPDGTEHGEFLALDLGGTNFRVLRVRVT 98
Query: 208 QN--EKPDIKETNYAIPKDKMTGKGEELFDYIAKTLSDFIKIHQIEKWPIKLGFTFSFPC 381
N ++ +++ YAIP+D M G G +LFD+IA+ L++F+ QI++ + LGFTFSFPC
Sbjct: 99 DNGLQRVEMENQIYAIPEDIMRGSGTQLFDHIAECLANFMDKLQIKEKKLPLGFTFSFPC 158
Query: 382 SQINLSKALLVRWTKGFTASDCEGQDVAALLQNAINKQ-NLNVKCVAVVNDTVGTLTSCA 558
Q L ++ LV WTKGF +S EG+DV L++ AI ++ + ++ VAVVNDTVGT+ +C
Sbjct: 159 HQTKLDESFLVSWTKGFKSSGVEGRDVVDLIRKAIQRRGDFDIDIVAVVNDTVGTMMTCG 218
Query: 559 LQDPKCGVGVIVGTGTNASYIEKIERVETFTEDKTGVEYVIINMEWGAFGDDGCLDKYRT 738
D C +G+IVGTG+NA Y+E++ ++ D+ + INMEWGAFGDDG L+ RT
Sbjct: 219 YDDQNCEIGLIVGTGSNACYMEEMRHIDMVEGDEGRM---CINMEWGAFGDDGTLNDIRT 275
Query: 739 KYDKLLDEQSLHPNQQIFEKMCSGMYLGEIVRLVLLDLIDRKILFRGEKPKSLEASGSFH 918
++D+ +D SL+P +Q+FEKM SGMY+GE+VRL+L+ + ++LF+G+ L +GSF
Sbjct: 276 EFDREIDMGSLNPGKQLFEKMISGMYMGELVRLILVKMAKAELLFQGKLSPELLTTGSFE 335
Query: 919 TKYLSEVERDPPHLLYCTSYMLTEDFNVPTFEEKDAAIVRAVCEKVAARAAFYIGTGIAC 1098
TK +S++E D + +Y + + +E D +C+ V+ R+A +A
Sbjct: 336 TKDVSDIEDDKDGIQ--KAYQILVRLGLSPLQE-DCVATHRICQIVSTRSASLCAATLAA 392
Query: 1099 LLNRI--NKKE----VTVGIDGSLFKFHCKFPERMTDIVHQLKNE-NIKFVLKLSEDGSG 1257
+L RI NK E T+G+DGS++K H F +R+ V +L + +++F+ SEDGSG
Sbjct: 393 VLWRIKENKGEERLRSTIGVDGSVYKKHPHFAKRLHKAVRRLVPDCDVRFL--RSEDGSG 450
Query: 1258 KG 1263
KG
Sbjct: 451 KG 452
>sp|P17710|HXK1_MOUSE Hexokinase type I (HK I) (Hexokinase, tumor isozyme) Length = 974 Score = 352 bits (902), Expect = 2e-96 Identities = 195/418 (46%), Positives = 267/418 (63%), Gaps = 10/418 (2%) Frame = +1 Query: 40 LEENMIRGLSAATHKDASVKMFPSYVTKLPTGKEEGQFLALDLGGTNYRVLLVTLRQNEK 219 L M GL T+ A+VKM PSYV +P G E G FLALDLGGTN+RVLLV +R +K Sbjct: 547 LRSEMEMGLRKETNSRATVKMLPSYVRSIPDGTEHGDFLALDLGGTNFRVLLVKIRSGKK 606 Query: 220 PDIKETN--YAIPKDKMTGKGEELFDYIAKTLSDFIKIHQIEKWPIKLGFTFSFPCSQIN 393 ++ N Y+IP + M G G+ELFD+I +SDF+ I+ + LGFTFSFPC Q + Sbjct: 607 RTVEMHNKIYSIPLEIMQGTGDELFDHIVSCISDFLDYMGIKGPRMPLGFTFSFPCKQTS 666 Query: 394 LSKALLVRWTKGFTASDCEGQDVAALLQNAINK-QNLNVKCVAVVNDTVGTLTSCALQDP 570 L +L+ WTKGF A+DC G DVA LL++A+ + + ++ VAVVNDTVGT+ +CA ++P Sbjct: 667 LDCGILITWTKGFKATDCVGHDVATLLRDAVKRREEFDLDVVAVVNDTVGTMMTCAYEEP 726 Query: 571 KCGVGVIVGTGTNASYIEKIERVETFTEDKTGVEYVIINMEWGAFGDDGCLDKYRTKYDK 750 C +G+IVGTG+NA Y+E+++ VE E G + INMEWGAFGD+GCLD RT +DK Sbjct: 727 SCEIGLIVGTGSNACYMEEMKNVE-MVEGNQG--QMCINMEWGAFGDNGCLDDIRTDFDK 783 Query: 751 LLDEQSLHPNQQIFEKMCSGMYLGEIVRLVLLDLIDRKILFRGEKPKSLEASGSFHTKYL 930 ++DE SL+ +Q FEKM SGMYLGEIVR +L+D + LFRG+ + L+ G F TK+L Sbjct: 784 VVDEYSLNSGKQRFEKMISGMYLGEIVRNILIDFTKKGFLFRGQISEPLKTRGIFETKFL 843 Query: 931 SEVERDPPHLLYCTSYMLTEDFNVPTFEEKDAAIVRAVCEKVAARAAFYIGTGIACLLNR 1110 S++E D LL + + N D+ +V+ VC V+ RAA G G+A ++ + Sbjct: 844 SQIESDRLALLQVRAILQQLGLNSTC---DDSILVKTVCGVVSKRAAQLCGAGMAAVVQK 900 Query: 1111 INKK------EVTVGIDGSLFKFHCKFPERMTDIVHQLKNE-NIKFVLKLSEDGSGKG 1263 I + VTVG+DG+L+K H F M V +L + + F+ LSEDGSGKG Sbjct: 901 IRENRGLDHLNVTVGVDGTLYKLHPHFSRIMHQTVKELSPKCTVSFL--LSEDGSGKG 956
Score = 299 bits (766), Expect = 1e-80
Identities = 166/423 (39%), Positives = 256/423 (60%), Gaps = 10/423 (2%)
Frame = +1
Query: 25 NIMHLLEENMIRGLSAATHKDASVKMFPSYVTKLPTGKEEGQFLALDLGGTNYRVLLVTL 204
+I+ ++ M GLS + ASVKM P++V +P G E+G F+ALDLGG+++R+L V +
Sbjct: 94 DILTRFKKEMKNGLSRDYNPTASVKMLPTFVRSIPDGSEKGDFIALDLGGSSFRILRVQV 153
Query: 205 RQNEKPDIKETN--YAIPKDKMTGKGEELFDYIAKTLSDFIKIHQIEKWPIKLGFTFSFP 378
+ ++ + Y P++ + G G +LFD++A+ L DF++ +I+ + +GFTFSFP
Sbjct: 154 NHEKSQNVSMESEVYDTPENIVHGSGSQLFDHVAECLGDFMEKRKIKDKKLPVGFTFSFP 213
Query: 379 CSQINLSKALLVRWTKGFTASDCEGQDVAALLQNAINKQ-NLNVKCVAVVNDTVGTLTSC 555
C Q + +A+L+ WTK F AS EG DV LL AI K+ + + VAVVNDTVGT+ +C
Sbjct: 214 CRQSKIDEAVLITWTKRFKASGVEGADVVKLLNKAIKKRGDYDANIVAVVNDTVGTMMTC 273
Query: 556 ALQDPKCGVGVIVGTGTNASYIEKIERVETFTEDKTGVEYVIINMEWGAFGDDGCLDKYR 735
D +C VG+I+GTGTNA Y+E++ ++ D+ + IN EWGAFGDDG L+ R
Sbjct: 274 GYDDQQCEVGLIIGTGTNACYMEELRHIDLVEGDEGRM---CINTEWGAFGDDGSLEDIR 330
Query: 736 TKYDKLLDEQSLHPNQQIFEKMCSGMYLGEIVRLVLLDLIDRKILFRGEKPKSLEASGSF 915
T++D+ LD SL+P +Q+FEKM SGMY+GE+VRL+L+ + +LF G L G F
Sbjct: 331 TEFDRELDRGSLNPGKQLFEKMVSGMYMGELVRLILVKMAKESLLFEGRITPELLTRGKF 390
Query: 916 HTKYLSEVERDPPHLLYCTSYMLTEDFNVPTFEEKDAAIVRAVCEKVAARAAFYIGTGIA 1095
T ++ +E D + +LT P+ + D V+ VC V+ R+A + +
Sbjct: 391 TTSDVAAIETDKEGVQNAKE-ILTRLGVEPSHD--DCVSVQHVCTIVSFRSANLVAATLG 447
Query: 1096 CLLNRIN------KKEVTVGIDGSLFKFHCKFPERMTDIVHQL-KNENIKFVLKLSEDGS 1254
+LNR+ + TVG+DGSL+K H ++ R + +L + +++F+ LSE GS
Sbjct: 448 AILNRLRDNKGTPRLRTTVGVDGSLYKMHPQYSRRFHKTLRRLVPDSDVRFL--LSESGS 505
Query: 1255 GKG 1263
GKG
Sbjct: 506 GKG 508
>sp|P27881|HXK2_RAT Hexokinase type II (HK II) Length = 917 Score = 351 bits (900), Expect = 3e-96 Identities = 193/418 (46%), Positives = 274/418 (65%), Gaps = 10/418 (2%) Frame = +1 Query: 40 LEENMIRGLSAATHKDASVKMFPSYVTKLPTGKEEGQFLALDLGGTNYRVLLVTLRQNEK 219 ++ M +GLS TH A VKM P+YV P G E+G FLALDLGGTN+RVLLV +R ++ Sbjct: 491 MKVEMEQGLSKETHAVAPVKMLPTYVCATPDGTEKGDFLALDLGGTNFRVLLVRVRNGKR 550 Query: 220 PDIKETN--YAIPKDKMTGKGEELFDYIAKTLSDFIKIHQIEKWPIKLGFTFSFPCSQIN 393 ++ N Y+IP++ M G GEELFD+I + ++DF++ ++ + LGFTFSFPC Q + Sbjct: 551 RGVEMHNKIYSIPQEVMHGTGEELFDHIVQCIADFLEYMGMKGVSLPLGFTFSFPCQQNS 610 Query: 394 LSKALLVRWTKGFTASDCEGQDVAALLQNAINK-QNLNVKCVAVVNDTVGTLTSCALQDP 570 L +++L++WTKGF AS CEG+DV LL+ AI++ + ++ VAVVNDTVGT+ +C +DP Sbjct: 611 LDQSILLKWTKGFKASGCEGEDVVTLLKEAIHRREEFDLDVVAVVNDTVGTMMTCGYEDP 670 Query: 571 KCGVGVIVGTGTNASYIEKIERVETFTEDKTGVEYVIINMEWGAFGDDGCLDKYRTKYDK 750 C VG+IVGTG+NA Y+E++ VE ++ + +NMEWGAFGD+GCLD RT +D Sbjct: 671 HCEVGLIVGTGSNACYMEEMRNVELVDGEEGRM---CVNMEWGAFGDNGCLDDLRTVFDV 727 Query: 751 LLDEQSLHPNQQIFEKMCSGMYLGEIVRLVLLDLIDRKILFRGEKPKSLEASGSFHTKYL 930 +DE SL+P +Q FEKM SGMYLGEIVR +L+D R +LFRG + L+ G F TK+L Sbjct: 728 AVDELSLNPGKQRFEKMISGMYLGEIVRNILIDFTKRGLLFRGRISERLKTRGIFETKFL 787 Query: 931 SEVERDPPHLLYCTSYMLTEDFNVPTFEEKDAAIVRAVCEKVAARAAFYIGTGIACLLNR 1110 S++E D LL + + D+ IV+ VC VA RAA G G+A ++++ Sbjct: 788 SQIESDCLALLQVRAILRHLGLESTC---DDSIIVKEVCTVVARRAAQLCGAGMAAVVDK 844 Query: 1111 INKK------EVTVGIDGSLFKFHCKFPERMTDIVHQLKNE-NIKFVLKLSEDGSGKG 1263 I + +VTVG+DG+L+K H F + M + V L + ++ F+ SEDGSGKG Sbjct: 845 IRENRGLDNLKVTVGVDGTLYKLHPHFAKVMHETVRDLAPKCDVSFL--ESEDGSGKG 900
Score = 325 bits (834), Expect = 1e-88
Identities = 179/422 (42%), Positives = 269/422 (63%), Gaps = 10/422 (2%)
Frame = +1
Query: 28 IMHLLEENMIRGLSAATHKDASVKMFPSYVTKLPTGKEEGQFLALDLGGTNYRVLLVTLR 207
I + M +GL A TH A+VKM P++V P G E G+FLALDLGGTN+RVL V +
Sbjct: 39 ISRRFRKEMEKGLGATTHPTAAVKMLPTFVRSTPDGTEHGEFLALDLGGTNFRVLRVRVT 98
Query: 208 QN--EKPDIKETNYAIPKDKMTGKGEELFDYIAKTLSDFIKIHQIEKWPIKLGFTFSFPC 381
N ++ +++ YAIP+D M G G +LFD+IA+ L++F+ QI++ + LGFTFSFPC
Sbjct: 99 DNGLQRVEMENQIYAIPEDIMRGSGTQLFDHIAECLANFMDKLQIKEKKLPLGFTFSFPC 158
Query: 382 SQINLSKALLVRWTKGFTASDCEGQDVAALLQNAINKQ-NLNVKCVAVVNDTVGTLTSCA 558
Q L ++ LV WTKGF +S EG+DV L++ AI ++ + ++ VAVVNDTVGT+ +C
Sbjct: 159 HQTKLDESFLVSWTKGFKSSGVEGRDVVDLIRKAIQRRGDFDIDIVAVVNDTVGTMMTCG 218
Query: 559 LQDPKCGVGVIVGTGTNASYIEKIERVETFTEDKTGVEYVIINMEWGAFGDDGCLDKYRT 738
D C +G+IVGTG+NA Y+E++ ++ D+ + INMEWGAFGDDG L+ RT
Sbjct: 219 YDDQNCEIGLIVGTGSNACYMEEMRHIDMVEGDEGRM---CINMEWGAFGDDGTLNDIRT 275
Query: 739 KYDKLLDEQSLHPNQQIFEKMCSGMYLGEIVRLVLLDLIDRKILFRGEKPKSLEASGSFH 918
++D+ +D SL+P +Q+FEKM SGMY+GE+VRL+L+ + ++LF+G+ L +GSF
Sbjct: 276 EFDREIDMGSLNPGKQLFEKMISGMYMGELVRLILVKMAKAELLFQGKLSPELLTTGSFE 335
Query: 919 TKYLSEVERDPPHLLYCTSYMLTEDFNVPTFEEKDAAIVRAVCEKVAARAAFYIGTGIAC 1098
TK +S++E D + ++ N ++D +C+ V+ R+A +A
Sbjct: 336 TKDVSDIEEDKDGIEKAYQILMRLGLNP---LQEDCVATHRICQIVSTRSASLCAATLAA 392
Query: 1099 LLNRI--NKKE----VTVGIDGSLFKFHCKFPERMTDIVHQLKNE-NIKFVLKLSEDGSG 1257
+L RI NK E T+G+DGS++K H F +R+ V +L + +++F+ SEDGSG
Sbjct: 393 VLWRIKENKGEERLRSTIGVDGSVYKKHPHFAKRLHKAVRRLVPDCDVRFL--RSEDGSG 450
Query: 1258 KG 1263
KG
Sbjct: 451 KG 452
>sp|Q9NFT7|HXK2_DROME Hexokinase type 2 Length = 486 Score = 347 bits (890), Expect = 5e-95 Identities = 180/414 (43%), Positives = 266/414 (64%), Gaps = 2/414 (0%) Frame = +1 Query: 28 IMHLLEENMIRGLSAATHKDASVKMFPSYVTKLPTGKEEGQFLALDLGGTNYRVLLVTLR 207 ++ + + + GL+ TH A +K F S+V LPTGKE G++LALDLGG+N+RVLLV L Sbjct: 59 VVERMTKEIKMGLAKDTHARAVIKCFVSHVQDLPTGKERGKYLALDLGGSNFRVLLVNLI 118 Query: 208 QNEKPDIKETNYAIPKDKMTGKGEELFDYIAKTLSDFIKIHQIEKWPIKLGFTFSFPCSQ 387 N + Y P+ M+G G+ LFD++A+ LS+F H +E + LGFTFSFP Q Sbjct: 119 SNSDVETMSKGYNFPQTLMSGSGKALFDFLAECLSEFCHSHGLENESLALGFTFSFPLQQ 178 Query: 388 INLSKALLVRWTKGFTASDCEGQDVAALLQNAINKQ-NLNVKCVAVVNDTVGTLTSCALQ 564 LSK +LV WTKGF+ G++V +LLQ AI+++ +L + VA++NDTVGTL SCA Sbjct: 179 QGLSKGILVAWTKGFSCEGVVGKNVVSLLQEAIDRRGDLKINTVAILNDTVGTLMSCAFY 238 Query: 565 DPKCGVGVIVGTGTNASYIEKIERVETFTEDKTGVE-YVIINMEWGAFGDDGCLDKYRTK 741 P C +G+IVGTG+NA Y+EK E F +T + +IIN EWGAFGD+G L+ RT Sbjct: 239 HPNCRIGLIVGTGSNACYVEKTVNAECFEGYQTSPKPSMIINCEWGAFGDNGVLEFVRTS 298 Query: 742 YDKLLDEQSLHPNQQIFEKMCSGMYLGEIVRLVLLDLIDRKILFRGEKPKSLEASGSFHT 921 YDK +D+ + +P +Q FEK SGMY+GE+VRLV+ D+I + +F G + ++ SF T Sbjct: 299 YDKAVDKVTPNPGKQTFEKCISGMYMGELVRLVITDMIAKGFMFHGIISEKIQERWSFKT 358 Query: 922 KYLSEVERDPPHLLYCTSYMLTEDFNVPTFEEKDAAIVRAVCEKVAARAAFYIGTGIACL 1101 Y+S+VE D P + +L+E + +E D +R +CE V++R+A G+ + Sbjct: 359 AYISDVESDAPGEYRNCNKVLSE-LGILGCQEPDKEALRYICEAVSSRSAKLCACGLVTI 417 Query: 1102 LNRINKKEVTVGIDGSLFKFHCKFPERMTDIVHQLKNENIKFVLKLSEDGSGKG 1263 +N++N EV +GIDGS+++FH K+ + + + +L +KF L +SEDGSG+G Sbjct: 418 INKMNINEVAIGIDGSVYRFHPKYHDMLQYHMKKLLKPGVKFELVVSEDGSGRG 471
>sp|P35557|HXK4_HUMAN Hexokinase D (Hexokinase type IV) (HK IV) (HK4) (Glucokinase) Length = 465 Score = 345 bits (884), Expect = 2e-94 Identities = 185/431 (42%), Positives = 268/431 (62%), Gaps = 11/431 (2%) Frame = +1 Query: 4 LKFNDYENIMHLLEENMIRGLSAATHKDASVKMFPSYVTKLPTGKEEGQFLALDLGGTNY 183 L+ D + +M +++ M RGL TH++ASVKM P+YV P G E G FL+LDLGGTN+ Sbjct: 25 LQEEDLKKVMRRMQKEMDRGLRLETHEEASVKMLPTYVRSTPEGSEVGDFLSLDLGGTNF 84 Query: 184 RVLLVTLRQNEKPD----IKETNYAIPKDKMTGKGEELFDYIAKTLSDFIKIHQIEKWPI 351 RV+LV + + E+ K Y+IP+D MTG E LFDYI++ +SDF+ HQ++ + Sbjct: 85 RVMLVKVGEGEEGQWSVKTKHQMYSIPEDAMTGTAEMLFDYISECISDFLDKHQMKHKKL 144 Query: 352 KLGFTFSFPCSQINLSKALLVRWTKGFTASDCEGQDVAALLQNAINKQ-NLNVKCVAVVN 528 LGFTFSFP ++ K +L+ WTKGF AS EG +V LL++AI ++ + + VA+VN Sbjct: 145 PLGFTFSFPVRHEDIDKGILLNWTKGFKASGAEGNNVVGLLRDAIKRRGDFEMDVVAMVN 204 Query: 529 DTVGTLTSCALQDPKCGVGVIVGTGTNASYIEKIERVETFTEDKTGVEYVIINMEWGAFG 708 DTV T+ SC +D +C VG+IVGTG NA Y+E+++ VE D+ + +N EWGAFG Sbjct: 205 DTVATMISCYYEDHQCEVGMIVGTGCNACYMEEMQNVELVEGDEGRM---CVNTEWGAFG 261 Query: 709 DDGCLDKYRTKYDKLLDEQSLHPNQQIFEKMCSGMYLGEIVRLVLLDLIDRKILFRGEKP 888 D G LD++ +YD+L+DE S +P QQ++EK+ G Y+GE+VRLVLL L+D +LF GE Sbjct: 262 DSGELDEFLLEYDRLVDESSANPGQQLYEKLIGGKYMGELVRLVLLRLVDENLLFHGEAS 321 Query: 889 KSLEASGSFHTKYLSEVERDPPHLLYCTSYMLTEDFNVPTFEEKDAAIVRAVCEKVAARA 1068 + L G+F T+++S+VE D + + T T D IVR CE V+ RA Sbjct: 322 EQLRTRGAFETRFVSQVESDTGDRKQIYNILSTLGLRPST---TDCDIVRRACESVSTRA 378 Query: 1069 AFYIGTGIACLLNRINKK------EVTVGIDGSLFKFHCKFPERMTDIVHQLKNENIKFV 1230 A G+A ++NR+ + +TVG+DGS++K H F ER V +L + + Sbjct: 379 AHMCSAGLAGVINRMRESRSEDVMRITVGVDGSVYKLHPSFKERFHASVRRL-TPSCEIT 437 Query: 1231 LKLSEDGSGKG 1263 SE+GSG+G Sbjct: 438 FIESEEGSGRG 448
>sp|P27595|HXK1_BOVIN Hexokinase type I (HK I) (Brain form hexokinase) Length = 918 Score = 345 bits (884), Expect = 2e-94 Identities = 189/418 (45%), Positives = 266/418 (63%), Gaps = 10/418 (2%) Frame = +1 Query: 40 LEENMIRGLSAATHKDASVKMFPSYVTKLPTGKEEGQFLALDLGGTNYRVLLVTLRQNEK 219 L M GL T+ +A+V M PS++ +P G E+G FLALDLGGTN+RVLLV +R +K Sbjct: 491 LRTEMEMGLRKETNSNATVNMLPSFLRSIPDGTEDGDFLALDLGGTNFRVLLVKIRSGKK 550 Query: 220 PDIKETN--YAIPKDKMTGKGEELFDYIAKTLSDFIKIHQIEKWPIKLGFTFSFPCSQIN 393 ++ N Y IP + M G GEELFD+I +SDF+ I+ + LGFTFSFPC Q + Sbjct: 551 STVEMHNKIYRIPIEIMQGTGEELFDHIVSCISDFLDYMGIKGPRMPLGFTFSFPCQQTS 610 Query: 394 LSKALLVRWTKGFTASDCEGQDVAALLQNAINK-QNLNVKCVAVVNDTVGTLTSCALQDP 570 L +L+ WTKGF A+DC G DV LL++A+ + + ++ VAVVNDTVGT+ +CA ++P Sbjct: 611 LDAGILITWTKGFKATDCVGHDVVTLLRDAVKRREEFDLDVVAVVNDTVGTMMTCAYEEP 670 Query: 571 KCGVGVIVGTGTNASYIEKIERVETFTEDKTGVEYVIINMEWGAFGDDGCLDKYRTKYDK 750 C VG+IVGTG+NA Y+E+++ VE ++ + INMEWGAFGD+GC D RT +DK Sbjct: 671 TCEVGLIVGTGSNACYMEEMKNVEMVEGNQ---RQMCINMEWGAFGDNGCSDDIRTDFDK 727 Query: 751 LLDEQSLHPNQQIFEKMCSGMYLGEIVRLVLLDLIDRKILFRGEKPKSLEASGSFHTKYL 930 ++DE SL+ Q FE M SG+YLGEIVR +L+D + LFRG+ + L+ G F TK+L Sbjct: 728 VVDEYSLNSGNQRFENMISGIYLGEIVRNILIDFTKKGFLFRGQISEPLKTRGIFETKFL 787 Query: 931 SEVERDPPHLLYCTSYMLTEDFNVPTFEEKDAAIVRAVCEKVAARAAFYIGTGIACLLNR 1110 S++E D LL + + N D+ +V+ VC V+ RAA G G+A ++ + Sbjct: 788 SQIESDRLALLQVRAILQQLGLNSTC---DDSILVKTVCGVVSKRAAQLCGAGMAAVVEK 844 Query: 1111 I------NKKEVTVGIDGSLFKFHCKFPERMTDIVHQLKNE-NIKFVLKLSEDGSGKG 1263 I ++ VTVG+DG+L+K H +F M V +L + N+ F+ LSEDGSGKG Sbjct: 845 IRENRGLDRLNVTVGVDGTLYKLHPQFSRIMHQTVKELSPKCNVSFL--LSEDGSGKG 900
Score = 272 bits (696), Expect = 1e-72
Identities = 159/423 (37%), Positives = 246/423 (58%), Gaps = 10/423 (2%)
Frame = +1
Query: 25 NIMHLLEENMIRGLSAATHKDASVKMFPSYVTKLPTGKEEGQFLALDLGGTNYRVLLVTL 204
+IM+ ++ M GLS + A+VKM P++V +P G E+G F+ALDLGG+++R+L V +
Sbjct: 38 DIMNRFKKEMKNGLSRDFNPTATVKMLPTFVRSIPDGSEKGDFIALDLGGSSFRILRVQV 97
Query: 205 R--QNEKPDIKETNYAIPKDKMTGKGEELFDYIAKTLSDFIKIHQIEKWPIKLGFTFSFP 378
QN ++ Y P++ M G G +LFD++ + L DF++ +I+ + +GFTFSFP
Sbjct: 98 NHEQNRPVHMESEVYDTPENIMHGSGSQLFDHVLECLGDFMEKKKIKDKKLPVGFTFSFP 157
Query: 379 CSQINLSKALLVRWTKGFTASDCEGQDVAALLQNAINKQ-NLNVKCVAVVNDTVGTLTSC 555
C Q + +A+L+ WTK F A EG V LL AI K+ + + VAVVNDTVGT+ C
Sbjct: 158 CRQSKIDQAILITWTKRFKARGAEGNYVVKLLDKAIKKRGDYDANIVAVVNDTVGTMIDC 217
Query: 556 ALQDPKCGVGVIVGTGTNASYIEKIERVETFTEDKTGVEYVIINMEWGAFGDDGCLDKYR 735
D C VG+I+GTGTNA Y+E++ +++ D + IN EWG GDDG L+ R
Sbjct: 218 GYDDQHCEVGLIIGTGTNACYMEELRQIDFGWGDD---GRMCINTEWGDLGDDGSLEDIR 274
Query: 736 TKYDKLLDEQSLHPNQQIFEKMCSGMYLGEIVRLVLLDLIDRKILFRGEKPKSLEASGSF 915
++D+ SL+P +Q FEKM SG Y+ ++VRLVL+ + +LF G L G F
Sbjct: 275 KEFDREFRRGSLNPGKQRFEKMVSGRYMEDVVRLVLVKMAKEGLLFEGRITPELLTRGKF 334
Query: 916 HTKYLSEVERDPPHLLYCTSYMLTEDFNVPTFEEKDAAIVRAVCEKVAARAAFYIGTGIA 1095
+T +S +E+D L+ +LT V ++ D V+ VC V+ R+A + +
Sbjct: 335 NTSDVSAIEKDKEG-LHNAKEILTR-LGVERSDD-DCVSVQHVCTIVSFRSANLVAATLG 391
Query: 1096 CLLNRIN------KKEVTVGIDGSLFKFHCKFPERMTDIVHQL-KNENIKFVLKLSEDGS 1254
+LNR+ + TV +DGSL+K H ++ R + +L + +++F+ LSE G+
Sbjct: 392 AILNRLRDNKSTPRLRTTVRVDGSLYKTHPQYSRRFHKTLRRLVPDSDVRFL--LSESGT 449
Query: 1255 GKG 1263
GKG
Sbjct: 450 GKG 452
>sp|P17712|HXK4_RAT Hexokinase D (Hexokinase type IV) (HK IV) (HK4) (Glucokinase) Length = 465 Score = 343 bits (880), Expect = 7e-94 Identities = 184/431 (42%), Positives = 266/431 (61%), Gaps = 11/431 (2%) Frame = +1 Query: 4 LKFNDYENIMHLLEENMIRGLSAATHKDASVKMFPSYVTKLPTGKEEGQFLALDLGGTNY 183 L+ D + +M +++ M RGL TH++ASVKM P+YV P G E G FL+LDLGGTN+ Sbjct: 25 LQEEDLKKVMSRMQKEMDRGLRLETHEEASVKMLPTYVRSTPEGSEVGDFLSLDLGGTNF 84 Query: 184 RVLLVTLRQNEKPD----IKETNYAIPKDKMTGKGEELFDYIAKTLSDFIKIHQIEKWPI 351 RV+LV + + E K Y+IP+D MTG E LFDYI++ +SDF+ HQ++ + Sbjct: 85 RVMLVKVGEGEAGQWSVKTKHQMYSIPEDAMTGTAEMLFDYISECISDFLDKHQMKHKKL 144 Query: 352 KLGFTFSFPCSQINLSKALLVRWTKGFTASDCEGQDVAALLQNAINKQ-NLNVKCVAVVN 528 LGFTFSFP +L K +L+ WTKGF AS EG ++ LL++AI ++ + + VA+VN Sbjct: 145 PLGFTFSFPVRHEDLDKGILLNWTKGFKASGAEGNNIVGLLRDAIKRRGDFEMDVVAMVN 204 Query: 529 DTVGTLTSCALQDPKCGVGVIVGTGTNASYIEKIERVETFTEDKTGVEYVIINMEWGAFG 708 DTV T+ SC +D +C VG+IVGTG NA Y+E+++ VE D+ + +N EWGAFG Sbjct: 205 DTVATMISCYYEDRQCEVGMIVGTGCNACYMEEMQNVELVEGDEGRM---CVNTEWGAFG 261 Query: 709 DDGCLDKYRTKYDKLLDEQSLHPNQQIFEKMCSGMYLGEIVRLVLLDLIDRKILFRGEKP 888 D G LD++ +YD+++DE S +P QQ++EK+ G Y+GE+VRLVLL L+D +LF GE Sbjct: 262 DSGELDEFLLEYDRMVDESSANPGQQLYEKIIGGKYMGELVRLVLLKLVDENLLFHGEAS 321 Query: 889 KSLEASGSFHTKYLSEVERDPPHLLYCTSYMLTEDFNVPTFEEKDAAIVRAVCEKVAARA 1068 + L G+F T+++S+VE D + + T D IVR CE V+ RA Sbjct: 322 EQLRTRGAFETRFVSQVESDSGDRKQIHNILSTLGLRPSV---TDCDIVRRACESVSTRA 378 Query: 1069 AFYIGTGIACLLNRINKK------EVTVGIDGSLFKFHCKFPERMTDIVHQLKNENIKFV 1230 A G+A ++NR+ + +TVG+DGS++K H F ER V +L N + Sbjct: 379 AHMCSAGLAGVINRMRESRSEDVMRITVGVDGSVYKLHPSFKERFHASVRRL-TPNCEIT 437 Query: 1231 LKLSEDGSGKG 1263 SE+GSG+G Sbjct: 438 FIESEEGSGRG 448
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 153,117,605
Number of Sequences: 369166
Number of extensions: 3170628
Number of successful extensions: 8947
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 8220
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 8731
length of database: 68,354,980
effective HSP length: 114
effective length of database: 47,295,190
effective search space used: 16647906880
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail