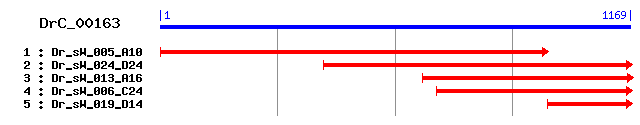
DrC_00163
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00163 (1168 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P55209|NP1L1_HUMAN Nucleosome assembly protein 1-like 1 ... 288 2e-77 sp|P28656|NP1L1_MOUSE Nucleosome assembly protein 1-like 1 ... 288 2e-77 sp|Q9Z2G8|NP1L1_RAT Nucleosome assembly protein 1-like 1 (N... 280 6e-75 sp|Q99733|NP1L4_HUMAN Nucleosome assembly protein 1-like 4 ... 270 4e-72 sp|P51860|NP1L2_MOUSE Nucleosome assembly protein 1-like 2 ... 193 9e-49 sp|Q9ULW6|NP1L2_HUMAN Nucleosome assembly protein 1-like 2 ... 191 3e-48 sp|O59797|YCO6_SCHPO Putative nucleosome assembly protein C... 173 1e-42 sp|P25293|NAP1_YEAST Nucleosome assembly protein 168 2e-41 sp|P78920|YB31_SCHPO Putative nucleosome assembly protein C... 167 7e-41 sp|Q99457|NP1L3_HUMAN Nucleosome assembly protein 1-like 3 115 3e-25
>sp|P55209|NP1L1_HUMAN Nucleosome assembly protein 1-like 1 (NAP-1 related protein) (hNRP) Length = 391 Score = 288 bits (738), Expect = 2e-77 Identities = 146/304 (48%), Positives = 204/304 (67%), Gaps = 2/304 (0%) Frame = +3 Query: 69 VLNNPEILASLQDEL-GLRMKAMKKFEQLPKNVKRRVRALRNLQLESIKIEESFFKELSV 245 ++ NP+ILA+LQ+ L GL E LP+ VKRRV AL+NLQ++ +IE F++E+ Sbjct: 42 MMQNPQILAALQERLDGLVETPTGYIESLPRVVKRRVNALKNLQVKCAQIEAKFYEEVHD 101 Query: 246 LEAKYSHIYQDVYDKRNEIVTGKHEPTDGECEYDSEAADELAEDVNDKAIISSGDEQIDN 425 LE KY+ +YQ ++DKR EI+ +EPT+ ECE+ + DE++E++ +KA I + + Sbjct: 102 LERKYAVLYQPLFDKRFEIINAIYEPTEEECEWKPDEEDEISEELKEKAKIEDEKKDEEK 161 Query: 426 NDVRGVPEFWLTVLENSELIRYNIQEHDKPILKHLIDMKCVLKNEDE-HGFVLEFYFSPN 602 D +G+PEFWLTV +N +L+ +QEHD+PILKHL D+K + + FVLEF+F PN Sbjct: 162 EDPKGIPEFWLTVFKNVDLLSDMVQEHDEPILKHLKDIKVKFSDAGQPMSFVLEFHFEPN 221 Query: 603 EFFSDEKLTKEYFVHVNAPTECPFEYEGPEIVRTVGCNIHWLPGKNVTVKLVKKVQKCKN 782 E+F++E LTK Y + PF ++GPEI+ GC I W GKNVT+K +KK QK K Sbjct: 222 EYFTNEVLTKTYRMRSEPDDSDPFSFDGPEIMGCTGCQIDWKKGKNVTLKTIKKKQKHKG 281 Query: 783 RSEKRTVTKVVKNDSFFNFFEPPTESLDGEDDIPSEIESLLESDFEMGQYIRERIIPRAV 962 R RTVTK V NDSFFNFF PP G D+ + E++L +DFE+G ++RERIIPR+V Sbjct: 282 RGTVRTVTKTVSNDSFFNFFAPPEVPESG--DLDDDAEAILAADFEIGHFLRERIIPRSV 339 Query: 963 LYFT 974 LYFT Sbjct: 340 LYFT 343
>sp|P28656|NP1L1_MOUSE Nucleosome assembly protein 1-like 1 (NAP-1 related protein) (Brain protein DN38) Length = 391 Score = 288 bits (737), Expect = 2e-77 Identities = 146/304 (48%), Positives = 205/304 (67%), Gaps = 2/304 (0%) Frame = +3 Query: 69 VLNNPEILASLQDEL-GLRMKAMKKFEQLPKNVKRRVRALRNLQLESIKIEESFFKELSV 245 ++ NP+ILA+LQ+ L GL E LPK VKRRV AL+NLQ++ +IE F++E+ Sbjct: 42 MMQNPQILAALQERLDGLVDTPTGYIESLPKVVKRRVNALKNLQVKCAQIEAKFYEEVHD 101 Query: 246 LEAKYSHIYQDVYDKRNEIVTGKHEPTDGECEYDSEAADELAEDVNDKAIISSGDEQIDN 425 LE KY+ +YQ ++DKR EI+ +EPT+ ECE+ + DE++E++ +KA I + + Sbjct: 102 LERKYAVLYQPLFDKRFEIINAIYEPTEEECEWKPDEEDEVSEELKEKAKIEDEKKDEEK 161 Query: 426 NDVRGVPEFWLTVLENSELIRYNIQEHDKPILKHLIDMKCVLKNEDE-HGFVLEFYFSPN 602 D +G+PEFWLTV +N +L+ +QEHD+PILKHL D+K + + FVLEF+F PN Sbjct: 162 EDPKGIPEFWLTVFKNVDLLSDMVQEHDEPILKHLKDIKVKFSDAGQPMSFVLEFHFEPN 221 Query: 603 EFFSDEKLTKEYFVHVNAPTECPFEYEGPEIVRTVGCNIHWLPGKNVTVKLVKKVQKCKN 782 ++F++E LTK Y + PF ++GPEI+ GC I W GKNVT+K +KK QK K Sbjct: 222 DYFTNEVLTKTYRMRSEPDDSDPFSFDGPEIMGCTGCQIDWKKGKNVTLKTIKKKQKHKG 281 Query: 783 RSEKRTVTKVVKNDSFFNFFEPPTESLDGEDDIPSEIESLLESDFEMGQYIRERIIPRAV 962 R RTVTK V NDSFFNFF PP +G D+ + E++L +DFE+G ++RERIIPR+V Sbjct: 282 RGTVRTVTKTVSNDSFFNFFAPPEVPENG--DLDDDAEAILAADFEIGHFLRERIIPRSV 339 Query: 963 LYFT 974 LYFT Sbjct: 340 LYFT 343
>sp|Q9Z2G8|NP1L1_RAT Nucleosome assembly protein 1-like 1 (NAP-1 related protein) Length = 390 Score = 280 bits (716), Expect = 6e-75 Identities = 144/304 (47%), Positives = 203/304 (66%), Gaps = 2/304 (0%) Frame = +3 Query: 69 VLNNPEILASLQDEL-GLRMKAMKKFEQLPKNVKRRVRALRNLQLESIKIEESFFKELSV 245 ++ NP+ILA+LQ+ L GL E LPK VKRRV AL+NLQ++ +IE F++E+ Sbjct: 42 MMQNPQILAALQERLDGLVDTPTGYIESLPKVVKRRVNALKNLQVKCAQIEAKFYEEVHD 101 Query: 246 LEAKYSHIYQDVYDKRNEIVTGKHEPTDGECEYDSEAADELAEDVNDKAIISSGDEQIDN 425 LE KY+ +YQ ++DKR EI+ +EPT+ ECE+ + DE++E++ +KA I + + Sbjct: 102 LERKYAVLYQPLFDKRFEIINAIYEPTEEECEWKPDEEDEVSEELKEKAKIEDEKKDEEK 161 Query: 426 NDVRGVPEFWLTVLENSELIRYNIQEHDKPILKHLIDMKCVLKNEDE-HGFVLEFYFSPN 602 D +G+PEFWLTV +N +L+ +QEHD+PILKHL D+K + + F+LEF+F PN Sbjct: 162 EDPKGIPEFWLTVFKN-DLLSDMVQEHDEPILKHLKDIKVKFSDAGQPMSFILEFHFEPN 220 Query: 603 EFFSDEKLTKEYFVHVNAPTECPFEYEGPEIVRTVGCNIHWLPGKNVTVKLVKKVQKCKN 782 E+F++E LTK Y + PF ++GPEI+ GC I W GKNVT+K +KK QK K Sbjct: 221 EYFTNEVLTKTYRMRSEPDDSDPFSFDGPEIMGCTGCQIDWKKGKNVTLKTIKKKQKHKG 280 Query: 783 RSEKRTVTKVVKNDSFFNFFEPPTESLDGEDDIPSEIESLLESDFEMGQYIRERIIPRAV 962 R RTVTK V SFFNFF PP +G D+ + E++L +DFE+G ++RERIIPR+V Sbjct: 281 RGTVRTVTKTVSKTSFFNFFAPPEVPENG--DLDDDXEAILAADFEIGHFLRERIIPRSV 338 Query: 963 LYFT 974 LYFT Sbjct: 339 LYFT 342
>sp|Q99733|NP1L4_HUMAN Nucleosome assembly protein 1-like 4 (Nucleosome assembly protein 2) (NAP2) Length = 375 Score = 270 bits (691), Expect = 4e-72 Identities = 145/324 (44%), Positives = 198/324 (61%), Gaps = 5/324 (1%) Frame = +3 Query: 18 SRHVAHKEKNDSSVSEDVLNNPEILASLQDEL-GLRMKAMKKFEQLPKNVKRRVRALRNL 194 S A N +++ V+ NP +LA+LQ+ L + E LPK VKRR+ AL+ L Sbjct: 14 SVEAAKNASNTEKLTDQVMQNPRVLAALQERLDNVPHTPSSYIETLPKAVKRRINALKQL 73 Query: 195 QLESIKIEESFFKELSVLEAKYSHIYQDVYDKRNEIVTGKHEPTDGECEYDSEAADE--L 368 Q+ IE F++E+ LE KY+ +YQ ++DKR E +TG EPTD E E+ SE +E L Sbjct: 74 QVRCAHIEAKFYEEVHDLERKYAALYQPLFDKRREFITGDVEPTDAESEWHSENEEEEKL 133 Query: 369 AEDVNDKAIISS-GDEQIDNNDVRGVPEFWLTVLENSELIRYNIQEHDKPILKHLIDMKC 545 A D+ K +++ + D +G+PEFW T+ N +++ +QE+D+PILKHL D+K Sbjct: 134 AGDMKSKVVVTEKAAATAEEPDPKGIPEFWFTIFRNVDMLSELVQEYDEPILKHLQDIKV 193 Query: 546 VLKNEDEH-GFVLEFYFSPNEFFSDEKLTKEYFVHVNAPTECPFEYEGPEIVRTVGCNIH 722 + + FVLEF+F PN++F++ LTK Y + PF +EGPEIV GC I Sbjct: 194 KFSDPGQPMSFVLEFHFEPNDYFTNSVLTKTYKMKSEPDKADPFSFEGPEIVDCDGCTID 253 Query: 723 WLPGKNVTVKLVKKVQKCKNRSEKRTVTKVVKNDSFFNFFEPPTESLDGEDDIPSEIESL 902 W GKNVTVK +KK QK K R RT+TK V N+SFFNFF P S DGE + + E Sbjct: 254 WKKGKNVTVKTIKKKQKHKGRGTVRTITKQVPNESFFNFFNPLKASGDGE-SLDEDSEFT 312 Query: 903 LESDFEMGQYIRERIIPRAVLYFT 974 L SDFE+G + RERI+PRAVLYFT Sbjct: 313 LASDFEIGHFFRERIVPRAVLYFT 336
>sp|P51860|NP1L2_MOUSE Nucleosome assembly protein 1-like 2 (Brain-specific protein, X-linked) Length = 460 Score = 193 bits (490), Expect = 9e-49 Identities = 112/317 (35%), Positives = 165/317 (52%), Gaps = 40/317 (12%) Frame = +3 Query: 144 EQLPKNVKRRVRALRNLQLESIKIEESFFKELSVLEAKYSHIYQDVYDKRNEIVTGKHEP 323 E LP VK RV AL+ LQ + +E F +E +E K++ +YQ + +KR +I+ +EP Sbjct: 102 ESLPVKVKCRVLALKKLQTRAAHLESKFLREFHDIERKFAEMYQPLLEKRRQIINAVYEP 161 Query: 324 TDGECEYDSEAADELAEDVN---------------------------------------D 386 T+ ECEY S+ D E+++ D Sbjct: 162 TEEECEYKSDCEDYFEEEMDEEEETNGNEDGMVHEYVDEDDGYEDCYYDYDDEEEEEEED 221 Query: 387 KAIISSGDEQIDNNDVRGVPEFWLTVLENSELIRYNIQEHDKPILKHLIDMKCVLKNEDE 566 + ++G E+++ D +G+P+FWLTVL+N E + I+++D+PILK L D+K L + E Sbjct: 222 DSAGATGGEEVNEEDPKGIPDFWLTVLKNVEALTPMIKKYDEPILKLLTDIKVKLSDPGE 281 Query: 567 H-GFVLEFYFSPNEFFSDEKLTKEYFVHVNAPTECPFEYEGPEIVRTVGCNIHWLPGKNV 743 F LEF+F PNE+F +E LTK Y + P Y G I GC+I W GKNV Sbjct: 282 PLSFTLEFHFKPNEYFKNELLTKTYVLKSKLACYDPHPYRGTAIEYATGCDIDWNEGKNV 341 Query: 744 TVKLVKKVQKCKNRSEKRTVTKVVKNDSFFNFFEPPTESLDGEDDIPSEIESLLESDFEM 923 T++ +KK Q+ + RTVT+ DSFFNFF P SL+G + DF + Sbjct: 342 TLRTIKKKQRHRVWGTVRTVTEDFPKDSFFNFFSPHGISLNG---------GVENDDFLL 392 Query: 924 GQYIRERIIPRAVLYFT 974 G +R IIPR+VL+F+ Sbjct: 393 GHNLRTYIIPRSVLFFS 409
>sp|Q9ULW6|NP1L2_HUMAN Nucleosome assembly protein 1-like 2 (Brain-specific protein, X-linked) Length = 460 Score = 191 bits (485), Expect = 3e-48 Identities = 113/317 (35%), Positives = 163/317 (51%), Gaps = 40/317 (12%) Frame = +3 Query: 144 EQLPKNVKRRVRALRNLQLESIKIEESFFKELSVLEAKYSHIYQDVYDKRNEIVTGKHEP 323 E LP VK RV AL+ LQ + +E F +E +E K++ +YQ + +KR +I+ +EP Sbjct: 102 ESLPVKVKYRVLALKKLQTRAANLESKFLREFHDIERKFAEMYQPLLEKRRQIINAIYEP 161 Query: 324 TDGECEYDSEAAD---------------------------------------ELAEDVND 386 T+ ECEY S++ D E E+ + Sbjct: 162 TEEECEYKSDSEDCDDEEMCHEEMYGNEEGMVHEYVDEDDGYEDYYYDYAVEEEEEEEEE 221 Query: 387 KAIISSGDEQIDNNDVRGVPEFWLTVLENSELIRYNIQEHDKPILKHLIDMKCVLKNEDE 566 I ++G+E + D +G+P+FWLTVL+N + + I+++D+PILK L D+K L + E Sbjct: 222 DDIEATGEENKEEEDPKGIPDFWLTVLKNVDTLTPLIKKYDEPILKLLTDIKVKLSDPGE 281 Query: 567 H-GFVLEFYFSPNEFFSDEKLTKEYFVHVNAPTECPFEYEGPEIVRTVGCNIHWLPGKNV 743 F LEF+F PNE+F +E LTK Y + P Y G I + GC I W GKNV Sbjct: 282 PLSFTLEFHFKPNEYFKNELLTKTYVLKSKLAYYDPHPYRGTAIEYSTGCEIDWNEGKNV 341 Query: 744 TVKLVKKVQKCKNRSEKRTVTKVVKNDSFFNFFEPPTESLDGEDDIPSEIESLLESDFEM 923 T+K +KK QK + RTVT+ DSFFNFF P + +G D DF + Sbjct: 342 TLKTIKKKQKHRIWGTIRTVTEDFPKDSFFNFFSPHGITSNGRDG---------NDDFLL 392 Query: 924 GQYIRERIIPRAVLYFT 974 G +R IIPR+VL+F+ Sbjct: 393 GHNLRTYIIPRSVLFFS 409
>sp|O59797|YCO6_SCHPO Putative nucleosome assembly protein C364.06 Length = 393 Score = 173 bits (438), Expect = 1e-42 Identities = 106/299 (35%), Positives = 165/299 (55%), Gaps = 2/299 (0%) Frame = +3 Query: 84 EILASLQDELGLRMKAMKK-FEQLPKNVKRRVRALRNLQLESIKIEESFFKELSVLEAKY 260 E L ++ +LG + + +LP+ V+RR+ LR LQ +E F KEL LE Y Sbjct: 48 ENLEAIDGKLGSLLHLTSEGVSELPEAVQRRISGLRGLQKRYSDLESQFQKELFELEKAY 107 Query: 261 SHIYQDVYDKRNEIVTGKHEPTDGECEYDSEAADELAEDVNDKAIISSGDEQIDNNDVRG 440 + Y ++ +R+E+V G EPT+ E + EAAD E+ + S +Q +D +G Sbjct: 108 AKKYAPIFKRRSEVVRGADEPTEEEIK-KGEAAD---ENEKKEPTSSESKKQEGGDDTKG 163 Query: 441 VPEFWLTVLENSELIRYNIQEHDKPILKHLIDMKCVLKNEDEHGFVLEFYFSPNEFFSDE 620 +PEFWLT ++N + I D+ L HL+D++ + ++ GF LEF F+ N FF+++ Sbjct: 164 IPEFWLTAMKNVLSLSEMITPEDEGALSHLVDIR--ISYMEKPGFKLEFEFAENPFFTNK 221 Query: 621 KLTKEYFVHVNAPTECPFEYEGPEIVRTVGCNIHWLPGKNVTVKLVKKVQKCKNRSEKRT 800 LTK Y+ + F Y+ E G + W ++TV+ V K Q+ KN + R Sbjct: 222 ILTKTYYYMEESGPSNVFLYDHAE-----GDKVDWKENADLTVRTVTKKQRNKNTKQTRV 276 Query: 801 VTKVVKNDSFFNFFEPPT-ESLDGEDDIPSEIESLLESDFEMGQYIRERIIPRAVLYFT 974 V V DSFFNFF PPT S + E+ E++ LLE D+++G+ +E++IPRAV +FT Sbjct: 277 VKVSVPRDSFFNFFNPPTPPSEEDEESESPELDELLELDYQIGEDFKEKLIPRAVEWFT 335
>sp|P25293|NAP1_YEAST Nucleosome assembly protein Length = 417 Score = 168 bits (426), Expect = 2e-41 Identities = 102/314 (32%), Positives = 173/314 (55%), Gaps = 10/314 (3%) Frame = +3 Query: 63 EDVL-NNPEILASLQDELGLRMKAMKKFEQ-LPKNVKRRVRALRNLQLESIKIEESFFKE 236 ED+L N P +L S+QD LG + + LPKNVK ++ +L+ LQ E ++E+ F E Sbjct: 57 EDILANQPLLLQSIQDRLGSLVGQDSGYVGGLPKNVKEKLLSLKTLQSELFEVEKEFQVE 116 Query: 237 LSVLEAKYSHIYQDVYDKRNEIVTGKHEPTDGECEYDSEAADELAEDVNDKAIISSGDEQ 416 + LE K+ Y+ ++++R+ I++G+ +P + E+ E +N+ ++ +E+ Sbjct: 117 MFELENKFLQKYKPIWEQRSRIISGQEQPKPEQI----AKGQEIVESLNETELLVDEEEK 172 Query: 417 IDNN----DVRGVPEFWLTVLENSELIRYNIQEHDKPILKHLIDMKCVLKNEDEHGFVLE 584 N+ V+G+P FWLT LEN ++ I + D +L++L D+ + GF L Sbjct: 173 AQNDSEEEQVKGIPSFWLTALENLPIVCDTITDRDAEVLEYLQDIGLEYLTDGRPGFKLL 232 Query: 585 FYF--SPNEFFSDEKLTKEYFVHVNAPTECPFEYEGPEIVRTVGCNIHWLP-GKNVTVKL 755 F F S N FF+++ L K YF F Y+ E GC I W NVTV L Sbjct: 233 FRFDSSANPFFTNDILCKTYFYQKELGYSGDFIYDHAE-----GCEISWKDNAHNVTVDL 287 Query: 756 VKKVQKCKNRSEKRTVTKVVKNDSFFNFFEPP-TESLDGEDDIPSEIESLLESDFEMGQY 932 + Q+ K + RT+ K+ +SFFNFF+PP ++ D ++++ ++E L D+ +G+ Sbjct: 288 EMRKQRNKTTKQVRTIEKITPIESFFNFFDPPKIQNEDQDEELEEDLEERLALDYSIGEQ 347 Query: 933 IRERIIPRAVLYFT 974 +++++IPRAV +FT Sbjct: 348 LKDKLIPRAVDWFT 361
>sp|P78920|YB31_SCHPO Putative nucleosome assembly protein C2D10.11C Length = 379 Score = 167 bits (422), Expect = 7e-41 Identities = 105/326 (32%), Positives = 167/326 (51%), Gaps = 8/326 (2%) Frame = +3 Query: 9 IKASRHVAHKEKNDSSVSEDVL----NNPEILASLQDELG-LRMKAMKKFEQLPKNVKRR 173 ++ +R + D S ++V NNP +L+ ++ L L K+ E L V+ R Sbjct: 35 LEKARQTLQSQLEDKSAHDEVSGLLRNNPAMLSMIEGRLSSLVGKSSGYIESLAPAVQNR 94 Query: 174 VRALRNLQLESIKIEESFFKELSVLEAKYSHIYQDVYDKRNEIVTGKHEPTDGECEYDSE 353 + AL+ LQ + I+ F +++ LE KY YQ ++ +R EI+ G EP D E +++ E Sbjct: 95 ITALKGLQKDCDAIQYEFRQKMLDLETKYEKKYQPIFSRRAEIIKGVSEPVDDELDHEEE 154 Query: 354 AADELAEDVNDKAIISSGDEQIDNN--DVRGVPEFWLTVLENSELIRYNIQEHDKPILKH 527 NN D +G+PEFWLT L N L+ I D+ +L+ Sbjct: 155 I--------------------FQNNLPDPKGIPEFWLTCLHNVFLVGEMITPEDENVLRS 194 Query: 528 LIDMKCVLKNEDEHGFVLEFYFSPNEFFSDEKLTKEYFVHVNAPTECPFEYEGPEIVRTV 707 L D++ + D HG+ LEF F N++F+++ LTK Y+ + F Y+ E Sbjct: 195 LSDIRFTNLSGDVHGYKLEFEFDSNDYFTNKILTKTYYYKDDLSPSGEFLYDHAE----- 249 Query: 708 GCNIHWL-PGKNVTVKLVKKVQKCKNRSEKRTVTKVVKNDSFFNFFEPPTESLDGEDDIP 884 G I+W+ P KN+TV++ K Q+ + ++ R V V NDSFFNFF PP D DD Sbjct: 250 GDKINWIKPEKNLTVRVETKKQRNRKTNQTRLVRTTVPNDSFFNFFSPPQLDDDESDDGL 309 Query: 885 SEIESLLESDFEMGQYIRERIIPRAV 962 + LLE D+++G+ +++IIP A+ Sbjct: 310 DDKTELLELDYQLGEVFKDQIIPLAI 335
>sp|Q99457|NP1L3_HUMAN Nucleosome assembly protein 1-like 3 Length = 506 Score = 115 bits (287), Expect = 3e-25 Identities = 65/183 (35%), Positives = 101/183 (55%), Gaps = 1/183 (0%) Frame = +3 Query: 429 DVRGVPEFWLTVLENSELIRYNIQEHDKPILKHLIDMKCVLKNEDEH-GFVLEFYFSPNE 605 D +G+P++WL VL+N + + IQ++D+PILK L D+ + + EF+F PN Sbjct: 307 DPKGIPDYWLIVLKNVDKLGPMIQKYDEPILKFLSDVSLKFSKPGQPVSYTFEFHFLPNP 366 Query: 606 FFSDEKLTKEYFVHVNAPTECPFEYEGPEIVRTVGCNIHWLPGKNVTVKLVKKVQKCKNR 785 +F +E L K Y + PF G EI GC I W GK+VTV + + Sbjct: 367 YFRNEVLVKTYIIKAKPDHNDPFFSWGWEIEDCKGCKIDWRRGKDVTVTTTQ--SRTTAT 424 Query: 786 SEKRTVTKVVKNDSFFNFFEPPTESLDGEDDIPSEIESLLESDFEMGQYIRERIIPRAVL 965 E +VV N SFFNFF PP + G+ + P E +++L+ DFE+GQ + + +I +++ Sbjct: 425 GEIEIQPRVVPNASFFNFFSPPEIPMIGKLE-pre-DAILDEDFEIGQILHDNVILKSIY 482 Query: 966 YFT 974 Y+T Sbjct: 483 YYT 485
Score = 69.7 bits (169), Expect = 2e-11
Identities = 33/82 (40%), Positives = 57/82 (69%), Gaps = 4/82 (4%)
Frame = +3
Query: 144 EQLPKNVKRRVRALRNLQLESIKIEESFFKELSVLEAKYSHIYQDVYDKRNEIVTGKHEP 323
++LP+ V+ RV+ALRN+Q E K++ F K + LE KY+ + + +YD+R +I+ ++EP
Sbjct: 98 DRLPQAVRNRVQALRNIQDECDKVDTLFLKAIHDLERKYAELNKPLYDRRFQIINAEYEP 157
Query: 324 TDGECEYDSE----AADELAED 377
T+ ECE++SE ++DE +D
Sbjct: 158 TEEECEWNSEDEEFSSDEEVQD 179
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 112,783,584
Number of Sequences: 369166
Number of extensions: 2062862
Number of successful extensions: 6740
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 6368
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6702
length of database: 68,354,980
effective HSP length: 113
effective length of database: 47,479,925
effective search space used: 13056979375
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail