DrC_00103
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
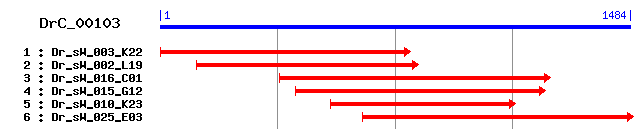
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00103 (1484 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q9NRA2|S17A5_HUMAN Sialin (Solute carrier family 17 memb... 367 e-101 sp|Q9MZD1|S17A5_SHEEP Sialin (Solute carrier family 17 memb... 358 2e-98 sp|Q8BN82|S17A5_MOUSE Sialin (Solute carrier family 17 memb... 357 4e-98 sp|Q9V7S5|PICO_DROME Putative inorganic phosphate cotranspo... 307 4e-83 sp|O61369|PICO_DROAN Putative inorganic phosphate cotranspo... 301 3e-81 sp|Q03567|YLD2_CAEEL Hypothetical protein C38C10.2 in chrom... 298 3e-80 sp|P34644|YOQ6_CAEEL Hypothetical protein ZK512.6 in chromo... 259 1e-68 sp|Q10046|YRT3_CAEEL Hypothetical protein T07A5.3 in chromo... 236 1e-61 sp|Q28722|NPT1_RABIT Renal sodium-dependent phosphate trans... 229 2e-59 sp|Q62795|NPT1_RAT Renal sodium-dependent phosphate transpo... 225 3e-58
>sp|Q9NRA2|S17A5_HUMAN Sialin (Solute carrier family 17 member 5) (Sodium/sialic acid cotransporter) (AST) (Membrane glycoprotein HP59) Length = 495 Score = 367 bits (943), Expect = e-101 Identities = 196/464 (42%), Positives = 280/464 (60%), Gaps = 6/464 (1%) Frame = +1 Query: 100 CIPTRYVISIWGFLGFAVVYVLRVNLSVAILAMVNSSALNEKGVNFTPECPVS----KSL 267 C RY ++I F GF +VY LRVNLSVA++ MV+S+ E + CP K Sbjct: 35 CCSARYNLAILAFFGFFIVYALRVNLSVALVDMVDSNTTLEDN-RTSKACPEHSAPIKVH 93 Query: 268 TNKTGNGEFVWSQEIQGIILGAFFWGYIPLQVPAGAIAVKIGGKWFFGIGVXXXXXXXXX 447 N+TG ++ W E QG ILG+FF+GYI Q+P G +A KIGGK G G+ Sbjct: 94 HNQTGK-KYQWDAETQGWILGSFFYGYIITQIPGGYVASKIGGKMLLGFGILGTAVLTLF 152 Query: 448 XXXAARAGYGWLIALRVLEGAGESITYPAMHTMLGYWAPAMETTFIGGIVYSGSFXXXXX 627 AA G G LI LR LEG GE +T+PAMH M WAP +E + + I Y+G+ Sbjct: 153 TPIAADLGVGPLIVLRALEGLGEGVTFPAMHAMWSSWAPPLERSKLLSISYAGAQLGTVI 212 Query: 628 XXXXXXXXXDANVMGGWPSVFYIPGIVGVLWFIIWSFVAYSTPAHHPRISEKEKRFIESS 807 W VFY G +G+ WF++W ++ TP H RIS EK +I SS Sbjct: 213 SLPLSGIIC---YYMNWTYVFYFFGTIGIFWFLLWIWLVSDTPQKHKRISHYEKEYILSS 269 Query: 808 VN--LSAEEKHTNVPWKQIMLSVPVWAISIANFCAMWSLYTLLTSLPQFFEYVLGFSISE 981 + LS+++ +VPW I+ S+P+WAI +A+F W+ YTLLT LP + + +L F++ E Sbjct: 270 LRNQLSSQK---SVPWVPILKSLPLWAIVVAHFSYNWTFYTLLTLLPTYMKEILRFNVQE 326 Query: 982 NGYINALPYFLSAITFVLSGKLADIIRSRKWMSTTVTRKLFNTIGMLVNAIFIVAISFIG 1161 NG++++LPY S + +LSG+ AD +R++ ST R++F+ IGM+ A+F+VA FIG Sbjct: 327 NGFLSSLPYLGSWLCMILSGQAADNLRAKWNFSTLCVRRIFSLIGMIGPAVFLVAAGFIG 386 Query: 1162 CDKVLVVTFLCIAVAVGTINSSGYNVNHIDLAPQYAGVLYGITNSIATVAGIIGPYVVGV 1341 CD L V FL I+ +G SSG+++NH+D+AP YAG+L GITN+ AT+ G++GP + Sbjct: 387 CDYSLAVAFLTISTTLGGFCSSGFSINHLDIAPSYAGILLGITNTFATIPGMVGPVIAKS 446 Query: 1342 LTNYNETKTQWQKVFYISSGITLFGAIFYLIFGSGDRQPWAIEN 1473 LT N T +WQ VFYI++ I +FGAIF+ +F G+ Q WA+ + Sbjct: 447 LTPDN-TVGEWQTVFYIAAAINVFGAIFFTLFAKGEVQNWALND 489
>sp|Q9MZD1|S17A5_SHEEP Sialin (Solute carrier family 17 member 5) (Sodium/sialic acid cotransporter) (Membrane glycoprotein SP55) Length = 495 Score = 358 bits (919), Expect = 2e-98 Identities = 197/463 (42%), Positives = 275/463 (59%), Gaps = 5/463 (1%) Frame = +1 Query: 100 CIPTRYVISIWGFLGFAVVYVLRVNLSVAILAMVNSSAL---NEKGVNFTPECPVSKSLT 270 C RY ++ F GF V+Y LRVNLSVA++ MV+S+ N K L Sbjct: 35 CCSARYNLAFLSFFGFFVLYSLRVNLSVALVDMVDSNTTAKDNRTSYECAEHSAPIKVLH 94 Query: 271 NKTGNGEFVWSQEIQGIILGAFFWGYIPLQVPAGAIAVKIGGKWFFGIGVXXXXXXXXXX 450 N+TG ++ W E QG ILG+FF+GYI Q+P G +A + GGK G G+ Sbjct: 95 NQTGK-KYRWDAETQGWILGSFFYGYIITQIPGGYVASRSGGKLLLGFGIFATAIFTLFT 153 Query: 451 XXAARAGYGWLIALRVLEGAGESITYPAMHTMLGYWAPAMETTFIGGIVYSGSFXXXXXX 630 AA G G L+ALR LEG GE +TYPAMH M WAP +E + + I Y+G+ Sbjct: 154 PLAADFGVGALVALRALEGLGEGVTYPAMHAMWSSWAPPLERSKLLSISYAGAQLGTVVS 213 Query: 631 XXXXXXXXDANVMGGWPSVFYIPGIVGVLWFIIWSFVAYSTPAHHPRISEKEKRFIESSV 810 W VFY GIVG++WFI+W + TP H I+ EK +I SS+ Sbjct: 214 LPLSGVIC---YYMNWTYVFYFFGIVGIIWFILWICLVSDTPETHKTITPYEKEYILSSL 270 Query: 811 N--LSAEEKHTNVPWKQIMLSVPVWAISIANFCAMWSLYTLLTSLPQFFEYVLGFSISEN 984 LS+++ +VPW ++ S+P+WAI +A+F W+ YTLLT LP + + VL F+I EN Sbjct: 271 KNQLSSQK---SVPWIPMLKSLPLWAIVVAHFSYNWTFYTLLTLLPTYMKEVLRFNIQEN 327 Query: 985 GYINALPYFLSAITFVLSGKLADIIRSRKWMSTTVTRKLFNTIGMLVNAIFIVAISFIGC 1164 G+++A+PY + +LSG+ AD +R+R ST R++F+ IGM+ AIF+VA FIGC Sbjct: 328 GFLSAVPYLGCWLCMILSGQAADNLRARWNFSTLWVRRVFSLIGMIGPAIFLVAAGFIGC 387 Query: 1165 DKVLVVTFLCIAVAVGTINSSGYNVNHIDLAPQYAGVLYGITNSIATVAGIIGPYVVGVL 1344 D L V FL I+ +G SSG+++NH+D+AP YAG+L GITN+ AT+ G+IGP + L Sbjct: 388 DYSLAVAFLTISTTLGGFCSSGFSINHLDIAPSYAGILLGITNTFATIPGMIGPIIARSL 447 Query: 1345 TNYNETKTQWQKVFYISSGITLFGAIFYLIFGSGDRQPWAIEN 1473 T N T +WQ VF I++ I +FGAIF+ +F G+ Q WAI + Sbjct: 448 TPEN-TIGEWQTVFCIAAAINVFGAIFFTLFAKGEVQNWAISD 489
>sp|Q8BN82|S17A5_MOUSE Sialin (Solute carrier family 17 member 5) (Sodium/sialic acid cotransporter) Length = 495 Score = 357 bits (917), Expect = 4e-98 Identities = 190/461 (41%), Positives = 269/461 (58%), Gaps = 3/461 (0%) Frame = +1 Query: 100 CIPTRYVISIWGFLGFAVVYVLRVNLSVAILAMVNSSAL---NEKGVNFTPECPVSKSLT 270 C RY ++I F GF V+Y LRVNLSVA++ MV+S+ N K Sbjct: 35 CCSARYNLAILAFCGFFVLYALRVNLSVALVDMVDSNTTLTDNRTSKECAEHSAPIKVHH 94 Query: 271 NKTGNGEFVWSQEIQGIILGAFFWGYIPLQVPAGAIAVKIGGKWFFGIGVXXXXXXXXXX 450 N TG ++ W E QG ILG+FF+GYI Q+P G IA ++GGK G+G+ Sbjct: 95 NHTGK-KYKWDAETQGWILGSFFYGYIVTQIPGGYIASRVGGKLLLGLGILGTSVFTLFT 153 Query: 451 XXAARAGYGWLIALRVLEGAGESITYPAMHTMLGYWAPAMETTFIGGIVYSGSFXXXXXX 630 AA G L+ LR LEG GE +T+PAMH M WAP +E + + I Y+G+ Sbjct: 154 PLAADLGVVTLVVLRALEGLGEGVTFPAMHAMWSSWAPPLERSKLLTISYAGAQLGTVIS 213 Query: 631 XXXXXXXXDANVMGGWPSVFYIPGIVGVLWFIIWSFVAYSTPAHHPRISEKEKRFIESSV 810 W VFY+ GIVG++WFI+W ++ TP H IS EK +I SS+ Sbjct: 214 LPLSGIIC---YYMNWTYVFYLFGIVGIVWFILWMWIVSDTPETHKTISHYEKEYIVSSL 270 Query: 811 NLSAEEKHTNVPWKQIMLSVPVWAISIANFCAMWSLYTLLTSLPQFFEYVLGFSISENGY 990 + VPW I+ S+P+WAI +A+F WS YTLLT LP + + +L F++ ENG+ Sbjct: 271 KNQLSSQKV-VPWGSILKSLPLWAIVVAHFSYNWSFYTLLTLLPTYMKEILRFNVQENGF 329 Query: 991 INALPYFLSAITFVLSGKLADIIRSRKWMSTTVTRKLFNTIGMLVNAIFIVAISFIGCDK 1170 ++ALPYF + +L G+ AD +R + ST R++F+ +GM+ A+F+VA FIGCD Sbjct: 330 LSALPYFGCWLCMILCGQAADYLRVKWNFSTISVRRIFSLVGMVGPAVFLVAAGFIGCDY 389 Query: 1171 VLVVTFLCIAVAVGTINSSGYNVNHIDLAPQYAGVLYGITNSIATVAGIIGPYVVGVLTN 1350 L V FL I+ +G SSG+++NH+D+AP YAG+L GITN+ AT+ G+ GP + LT Sbjct: 390 SLAVAFLTISTTLGGFASSGFSINHLDIAPSYAGILLGITNTFATIPGMTGPIIAKSLTP 449 Query: 1351 YNETKTQWQKVFYISSGITLFGAIFYLIFGSGDRQPWAIEN 1473 N T +WQ VF I++ I +FGAIF+ +F G+ Q WA+ + Sbjct: 450 DN-TIREWQTVFCIAAAINVFGAIFFTLFAKGEVQSWALSD 489
>sp|Q9V7S5|PICO_DROME Putative inorganic phosphate cotransporter Length = 529 Score = 307 bits (787), Expect = 4e-83 Identities = 169/458 (36%), Positives = 248/458 (54%), Gaps = 4/458 (0%) Frame = +1 Query: 100 CIPTRYVISIWGFLGFAVVYVLRVNLSVAILAMVNSSALNE-KGVNFTPECPVSKSLTNK 276 C TRY ++ FLG A YV+R N+SVAI+AMVN +A+ + + EC + Sbjct: 36 CFATRYFVTFMLFLGMANAYVMRTNMSVAIVAMVNHTAIKSGEAEEYDDECGDRDIPIDD 95 Query: 277 TGNGEFVWSQEIQGIILGAFFWGYIPLQVPAGAIAVKIGGKWFFGIGVXXXXXXXXXXXX 456 + +GEF WS +QG IL +FF+GY+ Q+P G +A K G F G G+ Sbjct: 96 SQDGEFAWSSALQGYILSSFFYGYVITQIPFGILAKKYGSLRFLGYGMLINSVFAFLVPV 155 Query: 457 AARAGYGW-LIALRVLEGAGESITYPAMHTMLGYWAPAMETTFIGGIVYSGSFXXXXXXX 633 AAR G W L A+R ++G GE P H ML W P E + +G VY+G+ Sbjct: 156 AARGGGVWGLCAVRFIQGLGEGPIVPCTHAMLAKWIPPNERSRMGAAVYAGAQFGTIISM 215 Query: 634 XXXXXXXDANVMGGWPSVFYIPGIVGVLWFIIWSFVAYSTPAHHPRISEKEKRFIESSVN 813 + GGWPS+FY+ GIVG +W I + + P+ HP I E+EK++I S+ Sbjct: 216 PLSGLLAEYGFDGGWPSIFYVFGIVGTVWSIAFLIFVHEDPSSHPTIDEREKKYINDSLW 275 Query: 814 LSAEEKHTNVPWKQIMLSVPVWAISIANFCAMWSLYTLLTSLPQFFEYVLGFSISENGYI 993 + K +P+K I+ S+P +AI A+ + TL+T LP + + VL FS+ NG + Sbjct: 276 GTDVVKSPPIPFKAIIKSLPFYAILFAHMGHNYGYETLMTELPTYMKQVLRFSLKSNGLL 335 Query: 994 NALPYFLSAITFVLSGKLADIIRSRKWMSTTVTRKLFNTIGMLVNAIFIVAISFIGCDKV 1173 ++LPY + + +AD + S K S T TRKL N+IG + ++A S+ GCD+ Sbjct: 336 SSLPYLAMWLFSMFISVVADWMISSKRFSHTATRKLINSIGQYGPGVALIAASYTGCDRA 395 Query: 1174 LVVTFLCIAVAVGTINSSGYNVNHIDLAPQYAGVLYGITNSIATVAGIIGPYVVGVLTN- 1350 L + L I V + SG+ +NH+DL P++AG L ITN A +AG++ P G L + Sbjct: 396 LTLAILTIGVGLNGGIYSGFKINHLDLTPRFAGFLMSITNCSANLAGLLAPIAAGHLISD 455 Query: 1351 -YNETKTQWQKVFYISSGITLFGAIFYLIFGSGDRQPW 1461 QWQ VF+I++ + + FY IFGSG+RQ W Sbjct: 456 PSKPMMGQWQIVFFIAAFVYIICGTFYNIFGSGERQYW 493
>sp|O61369|PICO_DROAN Putative inorganic phosphate cotransporter Length = 483 Score = 301 bits (771), Expect = 3e-81 Identities = 164/445 (36%), Positives = 242/445 (54%), Gaps = 3/445 (0%) Frame = +1 Query: 136 FLGFAVVYVLRVNLSVAILAMVNSSALNEKGVNFTPECPVSKSLTNKTGNGEFVWSQEIQ 315 FLG A YV+R N+SVAI+AMVN +A+ + EC + + +GEF W+ +Q Sbjct: 3 FLGMANAYVMRTNMSVAIVAMVNHTAIKSGEEEYDDECGDRDIPIDDSQDGEFPWNAALQ 62 Query: 316 GIILGAFFWGYIPLQVPAGAIAVKIGGKWFFGIGVXXXXXXXXXXXXAARAGYGW-LIAL 492 G IL +FF+GY+ Q+P G +A K G F G G+ AAR G W L A+ Sbjct: 63 GYILSSFFYGYVITQIPFGILAKKYGSLRFLGYGMLINSVFAFLVPVAAREGGVWGLCAV 122 Query: 493 RVLEGAGESITYPAMHTMLGYWAPAMETTFIGGIVYSGSFXXXXXXXXXXXXXXDANVMG 672 R ++G GE P H ML W P E + +G VY+G+ + G Sbjct: 123 RFIQGLGEGPIVPCTHAMLAKWIPPNERSRMGAAVYAGAQFGTIISMPLSGLLAEYGFDG 182 Query: 673 GWPSVFYIPGIVGVLWFIIWSFVAYSTPAHHPRISEKEKRFIESSVNLSAEEKHTNVPWK 852 GWPS+FY+ GIVG +W I + Y P+ HP+I E+EK++I S+ + K +P+K Sbjct: 183 GWPSIFYVFGIVGTVWSIAFLIFVYEDPSTHPKIDEREKKYINESLWGTDVIKSPPIPFK 242 Query: 853 QIMLSVPVWAISIANFCAMWSLYTLLTSLPQFFEYVLGFSISENGYINALPYFLSAITFV 1032 I+ S+P +AI A+ + TL+T LP + + VL FS+ NG +++LPY + + Sbjct: 243 SIVKSLPFYAILFAHMGHNYGYETLMTELPTYMKQVLRFSLKSNGLLSSLPYLAMWLLSM 302 Query: 1033 LSGKLADIIRSRKWMSTTVTRKLFNTIGMLVNAIFIVAISFIGCDKVLVVTFLCIAVAVG 1212 +AD + S K S T TRK+ N+IG + ++A S+ GCD+ L + L I V + Sbjct: 303 FISVIADWMISSKRFSLTATRKIINSIGQYGPGLALIAASYTGCDRALTLAILTIGVGLN 362 Query: 1213 TINSSGYNVNHIDLAPQYAGVLYGITNSIATVAGIIGPYVVGVLTN--YNETKTQWQKVF 1386 SG+ +NH+DL P++AG L ITN A +AG++ P G L + QWQ VF Sbjct: 363 GGIYSGFKINHLDLTPRFAGFLMSITNCSANLAGLLAPIAAGNLISDPSKPVMGQWQIVF 422 Query: 1387 YISSGITLFGAIFYLIFGSGDRQPW 1461 +I++ + + FY IFGSG+RQ W Sbjct: 423 FIAAFVYIICGTFYNIFGSGERQFW 447
>sp|Q03567|YLD2_CAEEL Hypothetical protein C38C10.2 in chromosome III Length = 493 Score = 298 bits (763), Expect = 3e-80 Identities = 162/460 (35%), Positives = 253/460 (55%), Gaps = 8/460 (1%) Frame = +1 Query: 109 TRYVISIWGFLGFAVVYVLRVNLSVAILAMVNSSALNEKGVNFTPECPVSKSLTNKTGN- 285 TR+ +S+ F G V Y++R N+S A++ MVN + + GV C K +T N Sbjct: 14 TRFALSLVMFFGCLVTYMMRTNMSFAVVCMVNENK-TDTGVEKVSRC--GKEMTPVESNS 70 Query: 286 ---GEFVWSQEIQGIILGAFFWGYIPLQVPAGAIAVKIGGKWFFGIGVXXXXXXXXXXXX 456 GEF W ++ G++L +FF+GYI Q+ G +A + GGK + + Sbjct: 71 SVIGEFDWDKQTTGMVLSSFFYGYIGSQIIGGHLASRYGGKRVVFVTILGSALLTLLNPV 130 Query: 457 AARAGYGWLIALRVLEGAGESITYPAMHTMLGYWAPAMETTFIGGIVYSGSFXXXXXXXX 636 AAR L LR G + T+PAMHTM W P +E + + G+ Y+G+ Sbjct: 131 AARTSEYALAILRAAIGFLQGATFPAMHTMWSVWGPPLELSVLTGVTYAGAQIGNVIVLP 190 Query: 637 XXXXXXDANVMGGWPSVFYIPGIVGVLWFIIWSFVAYSTPAHHPRISEKEKRFIESSVNL 816 + GGWPS+FYI G+ GVLW +W +V+ PA HPRI+ +EK++I ++V Sbjct: 191 LSGFLCEYGFDGGWPSIFYIIGVFGVLWTAVWWYVSSDKPATHPRITPEEKQYIVTAVEA 250 Query: 817 SAEE---KHTNVPWKQIMLSVPVWAISIANFCAMWSLYTLLTSLPQFFEYVLGFSISENG 987 S + K + PW +I+ S VWA +F W YT+L SLP F + VLG ++S G Sbjct: 251 SMGKDTGKVPSTPWIKILTSPAVWACWAGHFAGDWGAYTMLVSLPSFLKDVLGLNLSSLG 310 Query: 988 YINALPYFLSAITFVLSGKLADIIRSRKWMSTTVTRKLFNTIGMLVNAIFIVAISFIGCD 1167 + ++PY + G LAD +RS+ +ST TR+ + ++ IF+VA + GC Sbjct: 311 AVASIPYIAYFLAINAGGVLADTLRSKGILSTLNTRRAAMLVALIGQGIFLVASGYCGCG 370 Query: 1168 K-VLVVTFLCIAVAVGTINSSGYNVNHIDLAPQYAGVLYGITNSIATVAGIIGPYVVGVL 1344 + VLV+ F+ +A+ + +G+ VN++++AP ++G + G N+I+ +AGII P V L Sbjct: 371 QDVLVIIFITCGMAISGLQYAGFVVNYLEIAPPFSGTVMGTGNTISALAGIISPAVSSYL 430 Query: 1345 TNYNETKTQWQKVFYISSGITLFGAIFYLIFGSGDRQPWA 1464 T N T+ +WQ V ++++GI GA+ + IF SG+ QPWA Sbjct: 431 TP-NGTQEEWQMVLWLTAGILTIGALLFSIFASGEVQPWA 469
>sp|P34644|YOQ6_CAEEL Hypothetical protein ZK512.6 in chromosome III Length = 576 Score = 259 bits (663), Expect = 1e-68 Identities = 145/463 (31%), Positives = 237/463 (51%), Gaps = 3/463 (0%) Frame = +1 Query: 85 WYNRCCIPTRYVISIWGFLGFAVVYVLRVNLSVAILAMVNSSALNEKGVNFTPECPVSKS 264 W +C R++++I +GF + + +R N A M N+T Sbjct: 62 WIGKC--RKRWLLAILANMGFMISFGIRCNFGAAKTHMYK---------NYTDPY----- 105 Query: 265 LTNKTGNGEFVWSQEIQGIILGAFFWGYIPLQVPAGAIAVKIGGKWFFGIGVXXXXXXXX 444 K EF W+ + ++ ++F+GY+ Q+PAG +A K FG G+ Sbjct: 106 --GKVHMHEFNWTIDELSVMESSYFYGYLVTQIPAGFLAAKFPPNKLFGFGIGVGAFLNI 163 Query: 445 XXXXAARAGYGWLIA-LRVLEGAGESITYPAMHTMLGYWAPAMETTFIGGIVYSGSFXXX 621 + +L+A +++ +G + + YPAMH + YWAP ME + + ++GS+ Sbjct: 164 LLPYGFKVKSDYLVAFIQITQGLVQGVCYPAMHGVWRYWAPPMERSKLATTAFTGSYAGA 223 Query: 622 XXXXXXXXXXXDANVMGGWPSVFYIPGIVGVLWFIIWSFVAYSTPAHHPRISEKEKRFIE 801 W + FY+ G+ GV+W I+W V + PA HP IS++EK FIE Sbjct: 224 VLGLPLSAFLVS---YVSWAAPFYLYGVCGVIWAILWFCVTFEKPAFHPTISQEEKIFIE 280 Query: 802 SSVNLSAEEKHT--NVPWKQIMLSVPVWAISIANFCAMWSLYTLLTSLPQFFEYVLGFSI 975 ++ + T ++PWK I+ S PVWAI +ANF W+ Y LL + + + LG I Sbjct: 281 DAIGHVSNTHPTIRSIPWKAIVTSKPVWAIIVANFARSWTFYLLLQNQLTYMKEALGMKI 340 Query: 976 SENGYINALPYFLSAITFVLSGKLADIIRSRKWMSTTVTRKLFNTIGMLVNAIFIVAISF 1155 +++G + A+P+ + ++ G+LAD +RS K +STT RK+FN G A F++ +++ Sbjct: 341 ADSGLLAAIPHLVMGCVVLMGGQLADYLRSNKILSTTAVRKIFNCGGFGGEAAFMLIVAY 400 Query: 1156 IGCDKVLVVTFLCIAVAVGTINSSGYNVNHIDLAPQYAGVLYGITNSIATVAGIIGPYVV 1335 D ++ L AV + SG+NVNH+D+AP+YA +L G +N I T+AG+ P+V Sbjct: 401 TTSDTTAIMA-LIAAVGMSGFAISGFNVNHLDIAPRYAAILMGFSNGIGTLAGLTCPFVT 459 Query: 1336 GVLTNYNETKTQWQKVFYISSGITLFGAIFYLIFGSGDRQPWA 1464 T + +K W VF ++S I G FY ++ SG+ Q WA Sbjct: 460 EAFTAH--SKHGWTSVFLLASLIHFTGVTFYAVYASGELQEWA 500
>sp|Q10046|YRT3_CAEEL Hypothetical protein T07A5.3 in chromosome III Length = 544 Score = 236 bits (602), Expect = 1e-61 Identities = 146/458 (31%), Positives = 224/458 (48%), Gaps = 5/458 (1%) Frame = +1 Query: 112 RYVISIWGFLGFAVVYVLRVNLSVAILAMVNSSALNEKGVNFTPECPVSKSLTNKTGNGE 291 R+ I+I GFA+ + +R N VA MVN NFT + E Sbjct: 47 RWQIAILAHFGFAISFGIRSNFGVAKNRMVN---------NFT-------DAYGEVHERE 90 Query: 292 FVWSQEIQGIILGAFFWGYIPLQVPAGAIAVKIGGKWFFGIGVXXXXXXXXXXXXAARAG 471 F+W+ G++ +FF+GY Q+PAG +A K F +G+ + Sbjct: 91 FLWTGAEVGMMESSFFYGYAASQIPAGVLAAKFAPNKIFMLGILVASFMNILSAISFNFH 150 Query: 472 YG---WLIALRVLEGAGESITYPAMHTMLGYWAPAMETTFIGGIVYSGSFXXXXXXXXXX 642 +++ ++ ++G + YPAMH + +WAP +E + + ++GS Sbjct: 151 PYTDIFVMVVQAVQGLALGVLYPAMHGVWKFWAPPLERSKLATTAFTGSSVGVMTGLPAS 210 Query: 643 XXXXDANVMGGWPSVFYIPGIVGVLWFIIWSFVAYSTPAHHPRISEKEKRFIESSVNLSA 822 W + FY+ G+VG++W +IW +V+ +P H IS+ EK+ + + A Sbjct: 211 AYLVSHF---SWSTPFYVFGVVGIIWSLIWMYVSSHSPETHGYISDDEKKQVTEKIGDVA 267 Query: 823 EEKH--TNVPWKQIMLSVPVWAISIANFCAMWSLYTLLTSLPQFFEYVLGFSISENGYIN 996 + T +PW+ +M S VWAI I FC W + LL + + + VL I +G+I+ Sbjct: 268 VKNMSLTTLPWRDMMTSSAVWAIIICTFCRSWGFFLLLGNQLTYMKDVLHIDIKNSGFIS 327 Query: 997 ALPYFLSAITFVLSGKLADIIRSRKWMSTTVTRKLFNTIGMLVNAIFIVAISFIGCDKVL 1176 P F I + +G+L D +RS MST RK NT G V A+ + ++F+ D V+ Sbjct: 328 IFPQFGMCIVTLATGQLCDYLRSSGKMSTEAVRKSVNTFGFTVEAMMLGCLAFVR-DPVI 386 Query: 1177 VVTFLCIAVAVGTINSSGYNVNHIDLAPQYAGVLYGITNSIATVAGIIGPYVVGVLTNYN 1356 VT L IA SG+NVNH D+AP+YA +L GI N + VAG +G V +T N Sbjct: 387 AVTCLVIACTGSGSVLSGFNVNHFDIAPRYAPILMGIANGLGAVAG-VGGMVTNTVTYQN 445 Query: 1357 ETKTQWQKVFYISSGITLFGAIFYLIFGSGDRQPWAIE 1470 +W VF ++ I +FG IF+LIF GD PWA E Sbjct: 446 PDGWKW--VFLLAMAIDIFGVIFFLIFAKGDVLPWARE 481
>sp|Q28722|NPT1_RABIT Renal sodium-dependent phosphate transport protein 1 (Sodium/phosphate cotransporter 1) (Na(+)/PI cotransporter 1) (Renal sodium-phosphate transport protein 1) (Renal Na(+)-dependent phosphate cotransporter 1) (Solute carrier family 18 member 1) (NAPI-1) Length = 465 Score = 229 bits (583), Expect = 2e-59 Identities = 139/458 (30%), Positives = 220/458 (48%) Frame = +1 Query: 97 CCIPTRYVISIWGFLGFAVVYVLRVNLSVAILAMVNSSALNEKGVNFTPECPVSKSLTNK 276 C RYV++++ V+ R+ LS+ ++AMVN++ L+ +P K L N Sbjct: 12 CFCSFRYVLALFMHFCNIVIIAQRMCLSLTMVAMVNNTNLHG-----SPNTSAEKRLDN- 65 Query: 277 TGNGEFVWSQEIQGIILGAFFWGYIPLQVPAGAIAVKIGGKWFFGIGVXXXXXXXXXXXX 456 T N + WS ++QGII + F+G +Q+P G I+ K G + Sbjct: 66 TKNPVYNWSPDVQGIIFSSIFYGAFLIQIPVGYISGIYSIKKLIGFALFLSSLVSIFIPQ 125 Query: 457 AARAGYGWLIALRVLEGAGESITYPAMHTMLGYWAPAMETTFIGGIVYSGSFXXXXXXXX 636 AA G W+I RV++G + A H + WAP +E + + SG Sbjct: 126 AAAVGETWIIVCRVVQGITQGTVTTAQHEIWVKWAPPLERGRLTSMSLSGFLLGPFIVLL 185 Query: 637 XXXXXXDANVMGGWPSVFYIPGIVGVLWFIIWSFVAYSTPAHHPRISEKEKRFIESSVNL 816 ++ GWP VFYI G G ++W + Y P HP +S EK +I SS+ Sbjct: 186 VTGIICESL---GWPMVFYIFGACGCAVCLLWFVLYYDDPKDHPCVSLHEKEYITSSLIQ 242 Query: 817 SAEEKHTNVPWKQIMLSVPVWAISIANFCAMWSLYTLLTSLPQFFEYVLGFSISENGYIN 996 ++P K ++ S+P+WAIS F +W+ L+ P +L I ENG ++ Sbjct: 243 QGSSTRQSLPIKAMIKSLPLWAISFCCFAYLWTYSRLIVYTPTLINSMLHVDIRENGLLS 302 Query: 997 ALPYFLSAITFVLSGKLADIIRSRKWMSTTVTRKLFNTIGMLVNAIFIVAISFIGCDKVL 1176 +LPY + I V++G AD + SR +S T RKLF IG+L+ +F + + ++ Sbjct: 303 SLPYLFAWICGVIAGHTADFLMSRNMLSLTAIRKLFTAIGLLLPIVFSMCLLYLSSGFYS 362 Query: 1177 VVTFLCIAVAVGTINSSGYNVNHIDLAPQYAGVLYGITNSIATVAGIIGPYVVGVLTNYN 1356 +TFL +A A + G +N +DLAP+Y + G+T I G+ V G+ + Sbjct: 363 TITFLILANASSSFCLGGALINALDLAPRYYVFIKGVTTLIGMTGGMTSSTVAGLFLS-Q 421 Query: 1357 ETKTQWQKVFYISSGITLFGAIFYLIFGSGDRQPWAIE 1470 + ++ W K+F + S I + IFYLIF + Q WA E Sbjct: 422 DPESSWFKIFLLMSIINVISVIFYLIFAKAEIQDWAKE 459
>sp|Q62795|NPT1_RAT Renal sodium-dependent phosphate transport protein 1 (Sodium/phosphate cotransporter 1) (Na(+)/PI cotransporter 1) (Renal sodium-phosphate transport protein 1) (Renal Na(+)-dependent phosphate cotransporter 1) (Solute carrier family 18 member 1) Length = 465 Score = 225 bits (573), Expect = 3e-58 Identities = 136/456 (29%), Positives = 224/456 (49%), Gaps = 3/456 (0%) Frame = +1 Query: 112 RYVISIWGFLGFAVVYVLRVNLSVAILAMVNSSA---LNEKGVNFTPECPVSKSLTNKTG 282 RY ++I V+ RV L++ ++AMVN + L+ K V + + Sbjct: 17 RYGLAILLHFCNIVIMAQRVCLNLTMVAMVNKTEPPHLSNKSV---------AEMLDNVK 67 Query: 283 NGEFVWSQEIQGIILGAFFWGYIPLQVPAGAIAVKIGGKWFFGIGVXXXXXXXXXXXXAA 462 N WS +IQG++L + F G + +QVP G ++ + G + AA Sbjct: 68 NPVHSWSLDIQGLVLSSVFLGMVVIQVPVGYLSGAYPMEKIIGSSLFLSSVLSLLIPPAA 127 Query: 463 RAGYGWLIALRVLEGAGESITYPAMHTMLGYWAPAMETTFIGGIVYSGSFXXXXXXXXXX 642 + G +I RVL+G + H + WAP +E + + SG Sbjct: 128 QVGAALVIVCRVLQGIAQGAVSTGQHGIWVKWAPPLERGRLTSMTLSGFVMGPFIALLVS 187 Query: 643 XXXXDANVMGGWPSVFYIPGIVGVLWFIIWSFVAYSTPAHHPRISEKEKRFIESSVNLSA 822 D + GWP VFYI GIVG + + W + + P +HP +S EK +I SS+ Sbjct: 188 GFICD---LLGWPMVFYIFGIVGCVLSLFWFILLFDDPNNHPYMSSSEKDYITSSLMQQV 244 Query: 823 EEKHTNVPWKQIMLSVPVWAISIANFCAMWSLYTLLTSLPQFFEYVLGFSISENGYINAL 1002 ++P K ++ S+P+WAI + +F +WS L+T P F L ++ ENG +++L Sbjct: 245 HSGRQSLPIKAMLKSLPLWAIILNSFAFIWSNNLLVTYTPTFISTTLHVNVRENGLLSSL 304 Query: 1003 PYFLSAITFVLSGKLADIIRSRKWMSTTVTRKLFNTIGMLVNAIFIVAISFIGCDKVLVV 1182 PY L+ I +++G+++D + SRK S RKLF T+G+ IF+V + ++ + V Sbjct: 305 PYLLAYICGIVAGQMSDFLLSRKIFSVVAVRKLFTTLGIFCPVIFVVCLLYLSYNFYSTV 364 Query: 1183 TFLCIAVAVGTINSSGYNVNHIDLAPQYAGVLYGITNSIATVAGIIGPYVVGVLTNYNET 1362 FL +A + + + G +N +D+AP+Y G L +T I G+I + G++ N + Sbjct: 365 IFLTLANSTLSFSFCGQLINALDIAPRYYGFLKAVTALIGIFGGLISSTLAGLILN-QDP 423 Query: 1363 KTQWQKVFYISSGITLFGAIFYLIFGSGDRQPWAIE 1470 + W K F++ +GI + FYL+F GD Q WA E Sbjct: 424 EYAWHKNFFLMAGINVTCLAFYLLFAKGDIQDWAKE 459
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 181,965,738
Number of Sequences: 369166
Number of extensions: 4045341
Number of successful extensions: 11775
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 10717
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 11659
length of database: 68,354,980
effective HSP length: 115
effective length of database: 47,110,455
effective search space used: 17854862445
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail