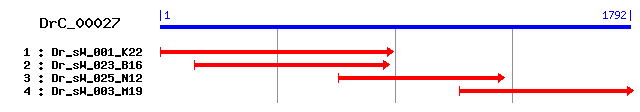
DrC_00027
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00027 (1792 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q29099|PTBP1_PIG Polypyrimidine tract-binding protein 1 ... 229 2e-59 sp|Q00438|PTBP1_RAT Polypyrimidine tract-binding protein 1 ... 229 3e-59 sp|Q8BHD7|ROD1_MOUSE Regulator of differentiation 1 (Rod1) 224 6e-58 sp|P26599|PTBP1_HUMAN Polypyrimidine tract-binding protein ... 146 2e-34 sp|Q9Z118|ROD1_RAT Regulator of differentiation 1 (Rod1) 131 6e-30 sp|O95758|ROD1_HUMAN Regulator of differentiation 1 (Rod1) 131 6e-30 sp|P17225|PTBP1_MOUSE Polypyrimidine tract-binding protein ... 119 2e-26 sp|Q8WVV9|HNRLL_HUMAN Heterogeneous nuclear ribonucleoprote... 107 9e-23 sp|Q921F4|HNRLL_MOUSE Heterogeneous nuclear ribonucleoprote... 107 1e-22 sp|P14866|HNRPL_HUMAN Heterogeneous nuclear ribonucleoprote... 71 1e-11
>sp|Q29099|PTBP1_PIG Polypyrimidine tract-binding protein 1 (PTB) (Heterogeneous nuclear ribonucleoprotein I) (hnRNP I) Length = 557 Score = 229 bits (584), Expect = 2e-59 Identities = 173/546 (31%), Positives = 260/546 (47%), Gaps = 26/546 (4%) Frame = +2 Query: 83 SDNEDSKKIRRE-----IPASKVLHIRSLPDDVTELEIASVATPFGHISYMVLTRKTGQA 247 ++ DSKK + + +P S+V+HIR LP DVTE E+ S+ PFG ++ +++ + QA Sbjct: 39 ANGNDSKKFKGDNRSAGVP-SRVIHIRKLPSDVTEGEVISLGLPFGKVTNLLMLKGKNQA 97 Query: 248 LLEMETKDIAETMFEYYQRHAPHLRNKRCFVQYXXXXXXXXXXXXXXPEAENAVTTANQI 427 +EM T++ A TM YY P LR + ++Q+ A+ A+ N + Sbjct: 98 FIEMNTEEAANTMVNYYTSVTPVLRGQPIYIQFSNHKELKTDSSPNQARAQAALQAVNSV 157 Query: 428 CD--------------SMAKESSGPVLRLVVDNWNHEKITNLHLFKIFQHYGILERIVIY 565 MA PVLR++V+N + +T L +IF +G + +I+ + Sbjct: 158 QSGNLALAASAAAVDAGMAMAGQSPVLRIIVENLFY-PVTLDVLHQIFSKFGTVLKIITF 216 Query: 566 VSKGVVQGFIQYSDPFSAKVAKALTNELCIFPRSCIMRVNFSNRQSLDISREDDLVRDFV 745 Q +QY+DP SA+ AK + I+ C +R++FS SL++ +D RD+ Sbjct: 217 TKNNQFQALLQYADPVSAQHAKLSLDGQNIYNACCTLRIDFSKLTSLNVKYNNDKSRDYT 276 Query: 746 RHPVSPQELKNVHVNIGGGVMKDVEXXXXXXXXXXXXXTXXXXXXXXXXXXNHPDDAMPL 925 R P P D + P A+P Sbjct: 277 R-PDLPS--------------GDNQPSLDQTMAAAFGAPGIMSASPYAGAGFPPTFAIPQ 321 Query: 926 LRSIA-PAIRGFLDNQDILSRDFSRNDRNDAYGRTHF----GEKTSPLLIVSNLNEAKIH 1090 +++ P + G L I S R A GR G S LL VSNLN ++ Sbjct: 322 AATVSVPNVHGALAPLAIP----SAAARAAAAGRIAIPGLAGAGNSVLL-VSNLNPERVT 376 Query: 1091 PDALFKLFGCYGDVTRVKIMYNRKNTALVQFANEEFAQRALQYLNGVNLYGKPLHVNFSK 1270 P +LF LFG Y DV RVKI++N+K ALVQ A+ AQ A+ +LNG L+GKP+ + SK Sbjct: 377 PQSLFILFGVYCDVQRVKILFNKKENALVQMADGSQAQLAMSHLNGHKLHGKPVRITLSK 436 Query: 1271 HDRISHRAPIAGSEEEASGDRLHKEYVDSKLHRFKRSQSRNHANIYEPSKVLHIAGLVDD 1450 H + + P G E++ L K+Y +S LHRFK+ S+N NI+ PS LH++ + Sbjct: 437 HQNV--QLpreGQEDQG----LTKDYGNSPLHRFKKPGSKNFQNIFPPSATLHLSNIPPS 490 Query: 1451 VSEEDIEALFQDNKFRVRKIRMFNSNGKKMXXXXXXXXXXXXXXXXXLHDYDLCGSN--L 1624 +SEED++ LF N V+ + F + +KM LH++DL G N L Sbjct: 491 ISEEDLKILFSSNGGIVKGFKFFQKD-RKMALIQMGSVEEAIQALIDLHNHDL-GENHHL 548 Query: 1625 RVSFSK 1642 RVSFSK Sbjct: 549 RVSFSK 554
>sp|Q00438|PTBP1_RAT Polypyrimidine tract-binding protein 1 (PTB) (Heterogeneous nuclear ribonucleoprotein I) (hnRNP I) (Pyrimidine-binding protein) (PYBP) Length = 555 Score = 229 bits (583), Expect = 3e-59 Identities = 172/554 (31%), Positives = 261/554 (47%), Gaps = 25/554 (4%) Frame = +2 Query: 56 FDAKEEETRSDNEDSKKIRRE-----IPASKVLHIRSLPDDVTELEIASVATPFGHISYM 220 F + ++ DSKK + + +P S+V+H+R LP DVTE E+ S+ PFG ++ + Sbjct: 29 FIMSSSASAANGNDSKKFKGDNRSTGVP-SRVIHVRKLPSDVTEGEVISLGLPFGKVTNL 87 Query: 221 VLTRKTGQALLEMETKDIAETMFEYYQRHAPHLRNKRCFVQYXXXXXXXXXXXXXXPEAE 400 ++ + QA +EM T++ A TM YY AP LR + ++Q+ A+ Sbjct: 88 LMLKGKNQAFIEMSTEEAANTMVNYYTSVAPVLRGQPIYIQFSNHKELKTDSSPNQARAQ 147 Query: 401 NAVTTANQICD--------------SMAKESSGPVLRLVVDNWNHEKITNLHLFKIFQHY 538 A+ N + MA PVLR++V+N + +T L +IF + Sbjct: 148 AALQAVNSVQSGNLALAASAAAVDAGMAMAGQSPVLRIIVENLFY-PVTLDVLHQIFSKF 206 Query: 539 GILERIVIYVSKGVVQGFIQYSDPFSAKVAKALTNELCIFPRSCIMRVNFSNRQSLDISR 718 G + +I+ + Q +QY+DP SA+ AK + I+ C +R++FS SL++ Sbjct: 207 GTVLKIITFTKNNQFQALLQYADPVSAQHAKLSLDGQNIYNACCTLRIDFSKLTSLNVKY 266 Query: 719 EDDLVRDFVRHPVSPQELKNVHVNIGGGVMKDVEXXXXXXXXXXXXXTXXXXXXXXXXXX 898 +D RD+ R P P G ++ + Sbjct: 267 NNDKSRDYTR-PDLP----------SGDSQPSLDQTMAAAFGAPGIMSASPYAGAVPSHL 315 Query: 899 NHPDDAMPLLRSIAPAIRGFLDNQDILSRDFSRNDRNDAYGRTHF----GEKTSPLLIVS 1066 HP A P + G L I S + A GR G S LL VS Sbjct: 316 CHPSRA----GLSVPNVHGALAPLAIPSAAAAA----AAAGRIAIPGLAGAGNSVLL-VS 366 Query: 1067 NLNEAKIHPDALFKLFGCYGDVTRVKIMYNRKNTALVQFANEEFAQRALQYLNGVNLYGK 1246 NLN ++ P +LF LFG YGDV RVKI++N+K ALV+ A+ AQ A+ +LNG L+GK Sbjct: 367 NLNPERVTPQSLFILFGVYGDVQRVKILFNKKENALVEMADGSQAQLAMSHLNGHKLHGK 426 Query: 1247 PLHVNFSKHDRISHRAPIAGSEEEASGDRLHKEYVDSKLHRFKRSQSRNHANIYEPSKVL 1426 + + SKH + + P G E++ L K+Y S LHRFK+ S+N NI+ PS L Sbjct: 427 SVRITLSKHQSV--QLpreGQEDQG----LTKDYGSSPLHRFKKPGSKNFQNIFPPSATL 480 Query: 1427 HIAGLVDDVSEEDIEALFQDNKFRVRKIRMFNSNGKKMXXXXXXXXXXXXXXXXXLHDYD 1606 H++ + VSE+D+++LF N V+ + F + +KM LH++D Sbjct: 481 HLSNIPPSVSEDDLKSLFSSNGGVVKGFKFFQKD-RKMALIQMGSVEEAVQALIELHNHD 539 Query: 1607 LCGSN--LRVSFSK 1642 L G N LRVSFSK Sbjct: 540 L-GENHHLRVSFSK 552
>sp|Q8BHD7|ROD1_MOUSE Regulator of differentiation 1 (Rod1) Length = 523 Score = 224 bits (571), Expect = 6e-58 Identities = 166/535 (31%), Positives = 259/535 (48%), Gaps = 15/535 (2%) Frame = +2 Query: 83 SDNEDSKKIRREIPASKVLHIRSLPDDVTELEIASVATPFGHISYMVLTRKTGQALLEME 262 +DN+ K R S+VLH+R +P DVTE E+ S+ PFG ++ +++ + QA LEM Sbjct: 14 NDNKKFKGDRPPCSPSRVLHLRKIPCDVTEAEVISLGLPFGKVTNLLMLKGKSQAFLEMA 73 Query: 263 TKDIAETMFEYYQRHAPHLRNKRCFVQYXXXXXXXXXXXXXXPEAENAVTTANQICD--- 433 +++ A TM YY PHLR++ ++QY A+ A+ + + Sbjct: 74 SEEAAVTMINYYTPVTPHLRSQPVYIQYSNHRELKTDNLPNQARAQAALQAVSAVQSGNL 133 Query: 434 ----SMAKESS-----GPVLRLVVDNWNHEKITNLHLFKIFQHYGILERIVIYVSKGVVQ 586 + A E + PVLR++++N + +T L +IF +G + +I+ + Q Sbjct: 134 SLPGATANEGTLLPGQSPVLRIIIENLFYP-VTLEVLHQIFSKFGTVLKIITFTKNNQFQ 192 Query: 587 GFIQYSDPFSAKVAKALTNELCIFPRSCIMRVNFSNRQSLDISREDDLVRDFVRHPVSPQ 766 +QY+DP +A+ AK + I+ C +R++FS SL++ +D RDF R Sbjct: 193 ALLQYADPVNAQYAKMALDGQNIYNACCTLRIDFSKLTSLNVKYNNDKSRDFTR------ 246 Query: 767 ELKNVHVNIGGGVMKDVEXXXXXXXXXXXXXTXXXXXXXXXXXXNHPDDAMPLLRSIA-P 943 + + G G +E + P A P ++ P Sbjct: 247 ----LDLPTGDG-QPSLEPPMAAAFGAPGIMSSPYAGAAGFA----PAIAFPQAAGLSVP 297 Query: 944 AIRGFLDNQDILSRDFSRNDRNDAYGRTHFGEKTSPLLIVSNLNEAKIHPDALFKLFGCY 1123 A+ G L + S S R G + G + +L+V+NLN I P LF LFG Y Sbjct: 298 AVPGALGPLTLTSSAVS--GRMAIPGAS--GMPGNSVLLVTNLNPDFITPHGLFILFGVY 353 Query: 1124 GDVTRVKIMYNRKNTALVQFANEEFAQRALQYLNGVNLYGKPLHVNFSKHDRISHRAPIA 1303 GDV RVKIM+N+K ALVQ A+ AQ A+ +L+G LYGK L SKH + + P Sbjct: 354 GDVHRVKIMFNKKENALVQMADASQAQLAMNHLSGQRLYGKVLRATLSKHQAV--QLpre 411 Query: 1304 GSEEEASGDRLHKEYVDSKLHRFKRSQSRNHANIYEPSKVLHIAGLVDDVSEEDIEALFQ 1483 G E++ L K++ +S LHRFK+ S+N NI+ PS LH++ + V+ +D++ LF Sbjct: 412 GQEDQG----LTKDFSNSPLHRFKKPGSKNFQNIFPPSATLHLSNIPPSVTMDDLKNLFT 467 Query: 1484 DNKFRVRKIRMFNSNGKKMXXXXXXXXXXXXXXXXXLHDYDLCGSN--LRVSFSK 1642 + V+ + F + +KM LH++DL G N LRVSFSK Sbjct: 468 EAGCSVKAFKFFQKD-RKMALIQLGSVEEAIQALIELHNHDL-GENHHLRVSFSK 520
>sp|P26599|PTBP1_HUMAN Polypyrimidine tract-binding protein 1 (PTB) (Heterogeneous nuclear ribonucleoprotein I) (hnRNP I) (57 kDa RNA-binding protein PPTB-1) Length = 531 Score = 146 bits (369), Expect = 2e-34 Identities = 85/199 (42%), Positives = 119/199 (59%), Gaps = 2/199 (1%) Frame = +2 Query: 1052 LLIVSNLNEAKIHPDALFKLFGCYGDVTRVKIMYNRKNTALVQFANEEFAQRALQYLNGV 1231 +L+VSNLN ++ P +LF LFG YGDV RVKI++N+K ALVQ A+ AQ A+ +LNG Sbjct: 338 VLLVSNLNPERVTPQSLFILFGVYGDVQRVKILFNKKENALVQMADGNQAQLAMSHLNGH 397 Query: 1232 NLYGKPLHVNFSKHDRISHRAPIAGSEEEASGDRLHKEYVDSKLHRFKRSQSRNHANIYE 1411 L+GKP+ + SKH + + P G E++ L K+Y +S LHRFK+ S+N NI+ Sbjct: 398 KLHGKPIRITLSKHQNV--QLpreGQEDQG----LTKDYGNSPLHRFKKPGSKNFQNIFP 451 Query: 1412 PSKVLHIAGLVDDVSEEDIEALFQDNKFRVRKIRMFNSNGKKMXXXXXXXXXXXXXXXXX 1591 PS LH++ + VSEED++ LF N V+ + F + +KM Sbjct: 452 PSATLHLSNIPPSVSEEDLKVLFSSNGGVVKGFKFFQKD-RKMALIQMGSVEEAVQALID 510 Query: 1592 LHDYDLCGSN--LRVSFSK 1642 LH++DL G N LRVSFSK Sbjct: 511 LHNHDL-GENHHLRVSFSK 528
Score = 117 bits (293), Expect = 1e-25
Identities = 71/240 (29%), Positives = 119/240 (49%), Gaps = 18/240 (7%)
Frame = +2
Query: 83 SDNEDSKKIRREIPA----SKVLHIRSLPDDVTELEIASVATPFGHISYMVLTRKTGQAL 250
++ DSKK + + + S+V+HIR LP DVTE E+ S+ PFG ++ +++ + QA
Sbjct: 39 ANGNDSKKFKGDSRSAGVPSRVIHIRKLPIDVTEGEVISLGLPFGKVTNLLMLKGKNQAF 98
Query: 251 LEMETKDIAETMFEYYQRHAPHLRNKRCFVQYXXXXXXXXXXXXXXPEAENAVTTANQIC 430
+EM T++ A TM YY P LR + ++Q+ A+ A+ N +
Sbjct: 99 IEMNTEEAANTMVNYYTSVTPVLRGQPIYIQFSNHKELKTDSSPNQARAQAALQAVNSVQ 158
Query: 431 D--------------SMAKESSGPVLRLVVDNWNHEKITNLHLFKIFQHYGILERIVIYV 568
MA PVLR++V+N + +T L +IF +G + +I+ +
Sbjct: 159 SGNLALAASAAAVDAGMAMAGQSPVLRIIVENLFY-PVTLDVLHQIFSKFGTVLKIITFT 217
Query: 569 SKGVVQGFIQYSDPFSAKVAKALTNELCIFPRSCIMRVNFSNRQSLDISREDDLVRDFVR 748
Q +QY+DP SA+ AK + I+ C +R++FS SL++ +D RD+ R
Sbjct: 218 KNNQFQALLQYADPVSAQHAKLSLDGQNIYNACCTLRIDFSKLTSLNVKYNNDKSRDYTR 277
Score = 34.3 bits (77), Expect = 1.2
Identities = 18/56 (32%), Positives = 29/56 (51%), Gaps = 2/56 (3%)
Frame = +2
Query: 1121 YGDVTRVKIMYNRKNTALVQFANEEFAQRALQYLNGVN--LYGKPLHVNFSKHDRI 1282
+G VT + +M KN A ++ EE A + Y V L G+P+++ FS H +
Sbjct: 82 FGKVTNL-LMLKGKNQAFIEMNTEEAANTMVNYYTSVTPVLRGQPIYIQFSNHKEL 136
Score = 33.9 bits (76), Expect = 1.6
Identities = 29/81 (35%), Positives = 45/81 (55%), Gaps = 6/81 (7%)
Frame = +2
Query: 1046 SPLL--IVSNLNEAKIHPDALFKLFGCYGDVTRVKIMYNRKNT--ALVQFANEEFAQRAL 1213
SP+L IV NL + D L ++F +G V ++ I + + N AL+Q+A+ AQ A
Sbjct: 181 SPVLRIIVENLFYP-VTLDVLHQIFSKFGTVLKI-ITFTKNNQFQALLQYADPVSAQHAK 238
Query: 1214 QYLNGVNLYGK--PLHVNFSK 1270
L+G N+Y L ++FSK
Sbjct: 239 LSLDGQNIYNACCTLRIDFSK 259
>sp|Q9Z118|ROD1_RAT Regulator of differentiation 1 (Rod1) Length = 523 Score = 131 bits (330), Expect = 6e-30 Identities = 81/199 (40%), Positives = 114/199 (57%), Gaps = 2/199 (1%) Frame = +2 Query: 1052 LLIVSNLNEAKIHPDALFKLFGCYGDVTRVKIMYNRKNTALVQFANEEFAQRALQYLNGV 1231 +L+V+NLN I P LF LFG YGDV RVKIM+N+K ALVQ A+ AQ A+ +L+G Sbjct: 330 VLLVTNLNPDFITPHGLFILFGVYGDVHRVKIMFNKKENALVQMADASQAQIAMNHLSGQ 389 Query: 1232 NLYGKPLHVNFSKHDRISHRAPIAGSEEEASGDRLHKEYVDSKLHRFKRSQSRNHANIYE 1411 LYGK L SKH + + P G E++ L K++ +S LHRFK+ S+N NI+ Sbjct: 390 RLYGKVLRATLSKHQAV--QLpreGQEDQG----LTKDFSNSPLHRFKKPGSKNFQNIFP 443 Query: 1412 PSKVLHIAGLVDDVSEEDIEALFQDNKFRVRKIRMFNSNGKKMXXXXXXXXXXXXXXXXX 1591 PS LH++ + V+ +D++ LF + V+ + F + +KM Sbjct: 444 PSATLHLSNIPPSVTMDDLKNLFTEAGCSVKAFKFFQKD-RKMALIQLGSVEEAIQALIE 502 Query: 1592 LHDYDLCGSN--LRVSFSK 1642 LH++DL G N LRVSFSK Sbjct: 503 LHNHDL-GENHHLRVSFSK 520
Score = 119 bits (298), Expect = 3e-26
Identities = 68/234 (29%), Positives = 117/234 (50%), Gaps = 12/234 (5%)
Frame = +2
Query: 83 SDNEDSKKIRREIPASKVLHIRSLPDDVTELEIASVATPFGHISYMVLTRKTGQALLEME 262
+DN+ K R S+VLH+R +P DVTE E+ S+ PFG ++ +++ + QA LEM
Sbjct: 14 NDNKKFKGDRPPCSPSRVLHLRKIPCDVTEAEVISLGLPFGKVTNLLMLKGKSQAFLEMA 73
Query: 263 TKDIAETMFEYYQRHAPHLRNKRCFVQYXXXXXXXXXXXXXXPEAENAVTTANQI----- 427
+++ A TM YY PHLR++ ++QY A+ A+ + +
Sbjct: 74 SEEAAVTMINYYTPVTPHLRSQPVYIQYSNHRELKTDNLPNQARAQAALQAVSAVQSGNL 133
Query: 428 -------CDSMAKESSGPVLRLVVDNWNHEKITNLHLFKIFQHYGILERIVIYVSKGVVQ 586
+ PVLR++++N + +T L +IF +G + +I+ + Q
Sbjct: 134 SLPGATSNEGTLLPGQSPVLRIIIENLFY-PVTLEVLHQIFSKFGTVLKIITFTKNNQFQ 192
Query: 587 GFIQYSDPFSAKVAKALTNELCIFPRSCIMRVNFSNRQSLDISREDDLVRDFVR 748
+QY+DP +A+ AK + I+ C +R++FS SL++ +D RDF R
Sbjct: 193 ALLQYADPVNAQYAKMALDGQNIYNACCTLRIDFSKLTSLNVKYNNDKSRDFTR 246
Score = 31.6 bits (70), Expect = 7.8
Identities = 21/63 (33%), Positives = 36/63 (57%), Gaps = 4/63 (6%)
Frame = +2
Query: 1094 DALFKLFGCYGDVTRVKIMYNRKNT--ALVQFANEEFAQRALQYLNGVNLYGK--PLHVN 1261
+ L ++F +G V ++ I + + N AL+Q+A+ AQ A L+G N+Y L ++
Sbjct: 167 EVLHQIFSKFGTVLKI-ITFTKNNQFQALLQYADPVNAQYAKMALDGQNIYNACCTLRID 225
Query: 1262 FSK 1270
FSK
Sbjct: 226 FSK 228
>sp|O95758|ROD1_HUMAN Regulator of differentiation 1 (Rod1) Length = 521 Score = 131 bits (330), Expect = 6e-30 Identities = 81/199 (40%), Positives = 114/199 (57%), Gaps = 2/199 (1%) Frame = +2 Query: 1052 LLIVSNLNEAKIHPDALFKLFGCYGDVTRVKIMYNRKNTALVQFANEEFAQRALQYLNGV 1231 +L+V+NLN I P LF LFG YGDV RVKIM+N+K ALVQ A+ AQ A+ +L+G Sbjct: 328 VLLVTNLNPDLITPHGLFILFGVYGDVHRVKIMFNKKENALVQMADANQAQLAMNHLSGQ 387 Query: 1232 NLYGKPLHVNFSKHDRISHRAPIAGSEEEASGDRLHKEYVDSKLHRFKRSQSRNHANIYE 1411 LYGK L SKH + + P G E++ L K++ +S LHRFK+ S+N NI+ Sbjct: 388 RLYGKVLRATLSKHQAV--QLpreGQEDQG----LTKDFSNSPLHRFKKPGSKNFQNIFP 441 Query: 1412 PSKVLHIAGLVDDVSEEDIEALFQDNKFRVRKIRMFNSNGKKMXXXXXXXXXXXXXXXXX 1591 PS LH++ + V+ +D++ LF + V+ + F + +KM Sbjct: 442 PSATLHLSNIPPSVTVDDLKNLFIEAGCSVKAFKFFQKD-RKMALIQLGSVEEAIQALIE 500 Query: 1592 LHDYDLCGSN--LRVSFSK 1642 LH++DL G N LRVSFSK Sbjct: 501 LHNHDL-GENHHLRVSFSK 518
Score = 125 bits (313), Expect = 5e-28
Identities = 73/237 (30%), Positives = 121/237 (51%), Gaps = 15/237 (6%)
Frame = +2
Query: 83 SDNEDSKKIRREIPA---SKVLHIRSLPDDVTELEIASVATPFGHISYMVLTRKTGQALL 253
++ DSKK +R+ P S+VLH+R +P DVTE EI S+ PFG ++ +++ + QA L
Sbjct: 9 ANGNDSKKFKRDRPPCSPSRVLHLRKIPCDVTEAEIISLGLPFGKVTNLLMLKGKSQAFL 68
Query: 254 EMETKDIAETMFEYYQRHAPHLRNKRCFVQYXXXXXXXXXXXXXXPEAENAVTTANQICD 433
EM +++ A TM YY PHLR++ ++QY A+ A+ + +
Sbjct: 69 EMASEEAAVTMVNYYTPITPHLRSQPVYIQYSNHRELKTDNLPNQARAQAALQAVSAVQS 128
Query: 434 SMAKESSG------------PVLRLVVDNWNHEKITNLHLFKIFQHYGILERIVIYVSKG 577
S G PVLR++++N + +T L +IF +G + +I+ +
Sbjct: 129 GSLALSGGPSNEGTVLPGQSPVLRIIIENLFY-PVTLEVLHQIFSKFGTVLKIITFTKNN 187
Query: 578 VVQGFIQYSDPFSAKVAKALTNELCIFPRSCIMRVNFSNRQSLDISREDDLVRDFVR 748
Q +QY+DP +A AK + I+ C +R++FS SL++ +D RDF R
Sbjct: 188 QFQALLQYADPVNAHYAKMALDGQNIYNACCTLRIDFSKLTSLNVKYNNDKSRDFTR 244
>sp|P17225|PTBP1_MOUSE Polypyrimidine tract-binding protein 1 (PTB) (Heterogeneous nuclear ribonucleoprotein I) (hnRNP I) Length = 527 Score = 119 bits (299), Expect = 2e-26 Identities = 73/250 (29%), Positives = 123/250 (49%), Gaps = 19/250 (7%) Frame = +2 Query: 56 FDAKEEETRSDNEDSKKIRRE-----IPASKVLHIRSLPDDVTELEIASVATPFGHISYM 220 F + ++ DSKK + + +P S+V+H+R LP DVTE E+ S+ PFG ++ + Sbjct: 29 FIMSSSASAANGNDSKKFKGDNRSAGVP-SRVIHVRKLPSDVTEGEVISLGLPFGKVTNL 87 Query: 221 VLTRKTGQALLEMETKDIAETMFEYYQRHAPHLRNKRCFVQYXXXXXXXXXXXXXXPEAE 400 ++ + QA +EM T++ A TM YY AP LR + ++Q+ A+ Sbjct: 88 LMLKGKNQAFIEMNTEEAANTMVNYYTSVAPVLRGQPIYIQFSNHKELKTDSSPNQVRAQ 147 Query: 401 NAVTTANQICD--------------SMAKESSGPVLRLVVDNWNHEKITNLHLFKIFQHY 538 A+ N + MA PVLR++V+N + +T L +IF + Sbjct: 148 AALQAVNSVQSGNLALAASAAAVDAGMAMAGQSPVLRIIVENLFY-PVTLDVLHQIFSKF 206 Query: 539 GILERIVIYVSKGVVQGFIQYSDPFSAKVAKALTNELCIFPRSCIMRVNFSNRQSLDISR 718 G + +I+ + Q +QY+DP SA+ AK + I+ C +R++FS SL++ Sbjct: 207 GTVLKIITFTKNNQFQALLQYADPVSAQHAKLSLDGQNIYNACCTLRIDFSKLTSLNVKY 266 Query: 719 EDDLVRDFVR 748 +D RD+ R Sbjct: 267 NNDKSRDYTR 276
Score = 105 bits (261), Expect = 6e-22
Identities = 74/203 (36%), Positives = 106/203 (52%), Gaps = 6/203 (2%)
Frame = +2
Query: 1052 LLIVSNLNEAKIHPDALFKLFGCYGDVTRVKIMYNRKNTALVQFANEEFAQRALQYLNGV 1231
+L+VSNLN ++ P +LF LFG YGDV RVKI++N+K ALVQ A+ AQ
Sbjct: 336 VLLVSNLNPERVTPQSLFILFGVYGDVQRVKILFNKKENALVQMADGSQAQ--------- 386
Query: 1232 NLYGKPLHVNFSKHDRISH----RAPIAGSEEEASGDRLHKEYVDSKLHRFKRSQSRNHA 1399
G+P + + +H + A + E L K+Y S L RFK+ S+N
Sbjct: 387 --LGEPPERAQAAREVSAHYTVQASECAAAREGQEDQGLTKDYGSSPL-RFKKPGSKNFQ 443
Query: 1400 NIYEPSKVLHIAGLVDDVSEEDIEALFQDNKFRVRKIRMFNSNGKKMXXXXXXXXXXXXX 1579
NI+ PS LH++ + VSE+D+++LF N V+ + F + +KM
Sbjct: 444 NIFPPSATLHLSNIPPSVSEDDLKSLFSSNGGVVKGFKFFQKD-RKMALIQMGSVEEAVQ 502
Query: 1580 XXXXLHDYDLCGSN--LRVSFSK 1642
LH++DL G N LRVSFSK
Sbjct: 503 ALIELHNHDL-GENHHLRVSFSK 524
Score = 33.9 bits (76), Expect = 1.6
Identities = 29/81 (35%), Positives = 45/81 (55%), Gaps = 6/81 (7%)
Frame = +2
Query: 1046 SPLL--IVSNLNEAKIHPDALFKLFGCYGDVTRVKIMYNRKNT--ALVQFANEEFAQRAL 1213
SP+L IV NL + D L ++F +G V ++ I + + N AL+Q+A+ AQ A
Sbjct: 180 SPVLRIIVENLFYP-VTLDVLHQIFSKFGTVLKI-ITFTKNNQFQALLQYADPVSAQHAK 237
Query: 1214 QYLNGVNLYGK--PLHVNFSK 1270
L+G N+Y L ++FSK
Sbjct: 238 LSLDGQNIYNACCTLRIDFSK 258
Score = 33.5 bits (75), Expect = 2.1
Identities = 18/56 (32%), Positives = 29/56 (51%), Gaps = 2/56 (3%)
Frame = +2
Query: 1121 YGDVTRVKIMYNRKNTALVQFANEEFAQRALQYLNGVN--LYGKPLHVNFSKHDRI 1282
+G VT + +M KN A ++ EE A + Y V L G+P+++ FS H +
Sbjct: 81 FGKVTNL-LMLKGKNQAFIEMNTEEAANTMVNYYTSVAPVLRGQPIYIQFSNHKEL 135
>sp|Q8WVV9|HNRLL_HUMAN Heterogeneous nuclear ribonucleoprotein L-like (Stromal RNA-regulating factor) (BLOCK24 protein) Length = 542 Score = 107 bits (268), Expect = 9e-23 Identities = 104/467 (22%), Positives = 187/467 (40%), Gaps = 8/467 (1%) Frame = +2 Query: 116 EIPASKVLHIRSLPDDVTELEIASVATPFGHISYMVLTRKTGQALLEMETKDIAETMFEY 295 ++ S V+H+R L + V E ++ FG I Y+++ QAL+E E D A+ + Sbjct: 71 KVSVSPVVHVRGLCESVVEADLVEALEKFGTICYVMMMPFKRQALVEFENIDSAKECVTF 130 Query: 296 YQRHAPHLRNKRCFVQYXXXXXXXXXXXXXXPEAENAVTTANQICDSMAKESSGPVLRLV 475 ++ ++ F Y P N V + P+ + Sbjct: 131 AADEPVYIAGQQAFFNYSTSKRITRPGNTDDPSGGNKV---------LLLSIQNPLYPIT 181 Query: 476 VDNWNHEKITNLHLFKIFQHYGILERIVIYVSKGVVQGFIQYSDPFSAKVAKALTNELCI 655 VD L+ + G ++RIVI+ G +Q +++ A+ AKA N I Sbjct: 182 VD----------VLYTVCNPVGKVQRIVIFKRNG-IQAMVEFESVLCAQKAKAALNGADI 230 Query: 656 FPRSCIMRVNFSNRQSLDISREDDLVRDFVRHPVSPQELKNVHVNIGGGVMKDVEXXXXX 835 + C +++ ++ L++ R D+ D+ + + ++ G G + Sbjct: 231 YAGCCTLKIEYARPTRLNVIRNDNDSWDYTKPYLGRRDR-------GKGRQRQA------ 277 Query: 836 XXXXXXXXTXXXXXXXXXXXXNHPDDAM--------PLLRSIAPAIRGFLDNQDILSRDF 991 HP PLL + G D ++++ Sbjct: 278 ------------------ILGEHPSSFRHDGYGSHGPLLPLPSRYRMGSRDTPELVAYPL 319 Query: 992 SRNDRNDAYGRTHFGEKTSPLLIVSNLNEAKIHPDALFKLFGCYGDVTRVKIMYNRKNTA 1171 + + +G G + +++VS L++ K++ +F LF YG++ +VK M TA Sbjct: 320 PQASSSYMHG----GNPSGSVVMVSGLHQLKMNCSRVFNLFCLYGNIEKVKFMKTIPGTA 375 Query: 1172 LVQFANEEFAQRALQYLNGVNLYGKPLHVNFSKHDRISHRAPIAGSEEEASGDRLHKEYV 1351 LV+ +E +RA+ +LN V L+GK L+V SK H + E G +K++ Sbjct: 376 LVEMGDEYAVERAVTHLNNVKLFGKRLNVCVSK----QHSVVPSQIFELEDGTSSYKDFA 431 Query: 1352 DSKLHRFKRSQSRNHANIYEPSKVLHIAGLVDDVSEEDIEALFQDNK 1492 SK +RF + + I PS VLH + V+EE L D++ Sbjct: 432 MSKNNRFTSAGQASKNIIQPPSCVLHYYNVPLCVTEETFTKLCNDHE 478
>sp|Q921F4|HNRLL_MOUSE Heterogeneous nuclear ribonucleoprotein L-like Length = 591 Score = 107 bits (266), Expect = 1e-22 Identities = 104/467 (22%), Positives = 188/467 (40%), Gaps = 8/467 (1%) Frame = +2 Query: 116 EIPASKVLHIRSLPDDVTELEIASVATPFGHISYMVLTRKTGQALLEMETKDIAETMFEY 295 ++ S V+H+R L + V E ++ FG I Y+++ QAL+E E D A+ + Sbjct: 120 KVSVSPVVHVRGLCESVVEADLVEALEKFGTICYVMMMPFKRQALVEFENIDSAKECVTF 179 Query: 296 YQRHAPHLRNKRCFVQYXXXXXXXXXXXXXXPEAENAVTTANQICDSMAKESSGPVLRLV 475 ++ ++ F Y P N V + P+ + Sbjct: 180 AADVPVYIAGQQAFFNYSTSKRITRPGNTDDPSGGNKV---------LLLSIQNPLYPIT 230 Query: 476 VDNWNHEKITNLHLFKIFQHYGILERIVIYVSKGVVQGFIQYSDPFSAKVAKALTNELCI 655 VD L+ + G ++RIVI+ G +Q +++ A+ AKA N I Sbjct: 231 VD----------VLYTVCNPVGKVQRIVIFKRNG-IQAMVEFESVLCAQKAKAALNGADI 279 Query: 656 FPRSCIMRVNFSNRQSLDISREDDLVRDFVRHPVSPQELKNVHVNIGGGVMKDVEXXXXX 835 + C +++ ++ L++ R D+ D+ + + ++ G G + Sbjct: 280 YAGCCTLKIEYARPTRLNVIRNDNDSWDYTKPYLGRRDR-------GKGRQRQA------ 326 Query: 836 XXXXXXXXTXXXXXXXXXXXXNHPDDAM--------PLLRSIAPAIRGFLDNQDILSRDF 991 +HP PLL + G D ++++ Sbjct: 327 ------------------ILGDHPSSFRHDGYGSHGPLLPLPSRYRMGSRDTPELVAYPL 368 Query: 992 SRNDRNDAYGRTHFGEKTSPLLIVSNLNEAKIHPDALFKLFGCYGDVTRVKIMYNRKNTA 1171 + + +G G + +++VS L++ K++ +F LF YG++ +VK M TA Sbjct: 369 PQASSSYMHG----GSPSGSVVMVSGLHQLKMNCSRVFNLFCLYGNIEKVKFMKTIPGTA 424 Query: 1172 LVQFANEEFAQRALQYLNGVNLYGKPLHVNFSKHDRISHRAPIAGSEEEASGDRLHKEYV 1351 LV+ +E +RA+ +LN V L+GK L+V SK H + E G +K++ Sbjct: 425 LVEMGDEYAVERAVTHLNNVKLFGKRLNVCVSK----QHSVVPSQIFELEDGTSSYKDFA 480 Query: 1352 DSKLHRFKRSQSRNHANIYEPSKVLHIAGLVDDVSEEDIEALFQDNK 1492 SK +RF + + I PS VLH + V+EE L D++ Sbjct: 481 MSKNNRFTSAGQASKNIIQPPSCVLHYYNVPLCVTEETFTKLCNDHE 527
>sp|P14866|HNRPL_HUMAN Heterogeneous nuclear ribonucleoprotein L (hnRNP L) Length = 558 Score = 70.9 bits (172), Expect = 1e-11 Identities = 44/140 (31%), Positives = 77/140 (55%) Frame = +2 Query: 1046 SPLLIVSNLNEAKIHPDALFKLFGCYGDVTRVKIMYNRKNTALVQFANEEFAQRALQYLN 1225 SP+L+V L+++K++ D +F +F YG+V +VK M ++ A+V+ A+ RA+ +LN Sbjct: 350 SPVLMVYGLDQSKMNGDRVFNVFCLYGNVEKVKFMKSKPGAAMVEMADGYAVDRAITHLN 409 Query: 1226 GVNLYGKPLHVNFSKHDRISHRAPIAGSEEEASGDRLHKEYVDSKLHRFKRSQSRNHANI 1405 ++G+ L+V SK I G E+ G +K++ +S+ +RF + I Sbjct: 410 NNFMFGQKLNVCVSKQPAIM-PGQSYGLED---GSCSYKDFSESRNNRFSTPEQAAKNRI 465 Query: 1406 YEPSKVLHIAGLVDDVSEED 1465 PS VLH +V+EE+ Sbjct: 466 QHPSNVLHFFNAPLEVTEEN 485
Score = 67.4 bits (163), Expect = 1e-10
Identities = 60/218 (27%), Positives = 95/218 (43%), Gaps = 3/218 (1%)
Frame = +2
Query: 122 PASKVLHIRSLPDDVTELEIASVATPFGHISYMVLTRKTGQALLEMETKDIAETMFEYYQ 301
PAS V+HIR L D V E ++ FG ISY+V+ K QAL+E E A Y
Sbjct: 68 PASPVVHIRGLIDGVVEADLVEALQEFGPISYVVVMPKKRQALVEFEDVLGACNAVNYAA 127
Query: 302 RHAPHLRNKRCFVQYXXXXXXXXXXXXXXPEAENAVTTANQIC---DSMAKESSGPVLRL 472
+ ++ FV Y +T+ +I DS S VL
Sbjct: 128 DNQIYIAGHPAFVNY---------------------STSQKISRPGDSDDSRSVNSVLLF 166
Query: 473 VVDNWNHEKITNLHLFKIFQHYGILERIVIYVSKGVVQGFIQYSDPFSAKVAKALTNELC 652
+ N + T++ L+ I G ++RIVI+ G VQ +++ SA+ AKA N
Sbjct: 167 TILNPIYSITTDV-LYTICNPCGPVQRIVIFRKNG-VQAMVEFDSVQSAQRAKASLNGAD 224
Query: 653 IFPRSCIMRVNFSNRQSLDISREDDLVRDFVRHPVSPQ 766
I+ C +++ ++ L++ + D D+ +S Q
Sbjct: 225 IYSGCCTLKIEYAKPTRLNVFKNDQDTWDYTNPNLSGQ 262
Score = 36.6 bits (83), Expect = 0.24
Identities = 28/109 (25%), Positives = 49/109 (44%), Gaps = 11/109 (10%)
Frame = +2
Query: 986 DFSRNDRNDAYGRTHFGEKTSPLLIVSNLNEA-KIHPDALFKLFGCYGDVTRVKIMYNRK 1162
++S + + G + + +L+ + LN I D L+ + G V R+ I
Sbjct: 141 NYSTSQKISRPGDSDDSRSVNSVLLFTILNPIYSITTDVLYTICNPCGPVQRIVIFRKNG 200
Query: 1163 NTALVQFANEEFAQRALQYLNGVNL----------YGKPLHVNFSKHDR 1279
A+V+F + + AQRA LNG ++ Y KP +N K+D+
Sbjct: 201 VQAMVEFDSVQSAQRAKASLNGADIYSGCCTLKIEYAKPTRLNVFKNDQ 249
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 195,125,817
Number of Sequences: 369166
Number of extensions: 3914569
Number of successful extensions: 10204
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 9492
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 10178
length of database: 68,354,980
effective HSP length: 116
effective length of database: 46,925,720
effective search space used: 22524345600
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail