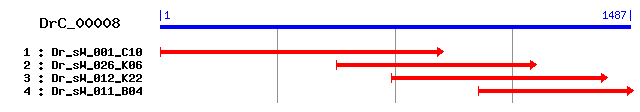
DrC_00008
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00008 (1487 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q9Y6E0|STK24_HUMAN Serine/threonine-protein kinase 24 (S... 185 3e-46 sp|Q99KH8|STK24_MOUSE Serine/threonine-protein kinase 24 184 8e-46 sp|Q99JT2|MST4_MOUSE Serine/threonine-protein kinase MST4 (... 177 9e-44 sp|Q9P289|MST4_HUMAN Serine/threonine-protein kinase MST4 (... 176 1e-43 sp|O22040|M3K1_ARATH Mitogen-activated protein kinase kinas... 175 3e-43 sp|Q9VXE5|PAKM_DROME Serine/threonine-protein kinase PAK mb... 173 1e-42 sp|O22042|M3K3_ARATH Mitogen-activated protein kinase kinas... 173 1e-42 sp|Q9FZ36|M3K2_ARATH Mitogen-activated protein kinase kinas... 172 2e-42 sp|Q13177|PAK2_HUMAN Serine/threonine-protein kinase PAK 2 ... 172 2e-42 sp|Q13043|STK4_HUMAN Serine/threonine-protein kinase 4 (STE... 171 4e-42
>sp|Q9Y6E0|STK24_HUMAN Serine/threonine-protein kinase 24 (STE20-like kinase MST3) (MST-3) (Mammalian STE20-like protein kinase 3) Length = 443 Score = 185 bits (469), Expect = 3e-46 Identities = 105/275 (38%), Positives = 167/275 (60%), Gaps = 3/275 (1%) Frame = +3 Query: 591 NVEPDFENL-VKLEKLGKGSFGTVFKCVSIETKGEMAVKKLKIPTTDKTKEEKLSVEREL 767 N++ D E L KLEK+GKGSFG VFK + T+ +A+K + + ++ ++E +++E+ Sbjct: 27 NLKADPEELFTKLEKIGKGSFGEVFKGIDNRTQKVVAIKIIDL---EEAEDEIEDIQQEI 83 Query: 768 QVYSKHSDHPRIVKYERNFKTEDAIFLVMEYMKGGSLGGYIRKNGPLSENKIKKYSKQIL 947 V S+ D P + KY ++ + ++++MEY+ GGS + GPL E +I ++IL Sbjct: 84 TVLSQ-CDSPYVTKYYGSYLKDTKLWIIMEYLGGGSALDLLEP-GPLDETQIATILREIL 141 Query: 948 EALQYLHEQKILHRDIKSDNILLDEDDNVKLCDFGLSKILTDLAAKDFLNGAIQSSVGTP 1127 + L YLH +K +HRDIK+ N+LL E VKL DFG++ LTD K + VGTP Sbjct: 142 KGLDYLHSEKKIHRDIKAANVLLSEHGEVKLADFGVAGQLTDTQIKR------NTFVGTP 195 Query: 1128 NYMAPEIMNEEPYGSSADVWSFGATMVEMVTGFPPYHQMGVCKVMKIISESR--KIEYNY 1301 +MAPE++ + Y S AD+WS G T +E+ G PP+ ++ KV+ +I ++ +E NY Sbjct: 196 FWMAPEVIKQSAYDSKADIWSLGITAIELARGEPPHSELHPMKVLFLIPKNNPPTLEGNY 255 Query: 1302 PNNLSDNLKSFIDSCLQSEPARRLTSKQLMNHPFL 1406 S LK F+++CL EP+ R T+K+L+ H F+ Sbjct: 256 ----SKPLKEFVEACLNKEPSFRPTAKELLKHKFI 286
>sp|Q99KH8|STK24_MOUSE Serine/threonine-protein kinase 24 Length = 431 Score = 184 bits (466), Expect = 8e-46 Identities = 105/275 (38%), Positives = 167/275 (60%), Gaps = 3/275 (1%) Frame = +3 Query: 591 NVEPDFENL-VKLEKLGKGSFGTVFKCVSIETKGEMAVKKLKIPTTDKTKEEKLSVEREL 767 N++ D E L KLEK+GKGSFG VFK + T+ +A+K + + ++ ++E +++E+ Sbjct: 15 NLKADPEELFTKLEKIGKGSFGEVFKGIDNRTQKVVAIKIIDL---EEAEDEIEDIQQEI 71 Query: 768 QVYSKHSDHPRIVKYERNFKTEDAIFLVMEYMKGGSLGGYIRKNGPLSENKIKKYSKQIL 947 V S+ D P + KY ++ + ++++MEY+ GGS + GPL E +I ++IL Sbjct: 72 TVLSQ-CDSPYVTKYYGSYLKDTKLWIIMEYLGGGSALDLLEP-GPLDEIQIATILREIL 129 Query: 948 EALQYLHEQKILHRDIKSDNILLDEDDNVKLCDFGLSKILTDLAAKDFLNGAIQSSVGTP 1127 + L YLH +K +HRDIK+ N+LL E VKL DFG++ LTD K + VGTP Sbjct: 130 KGLDYLHSEKKIHRDIKAANVLLSEHGEVKLADFGVAGQLTDTQIKR------NTFVGTP 183 Query: 1128 NYMAPEIMNEEPYGSSADVWSFGATMVEMVTGFPPYHQMGVCKVMKIISESR--KIEYNY 1301 +MAPE++ + Y S AD+WS G T +E+ G PP+ ++ KV+ +I ++ +E NY Sbjct: 184 FWMAPEVIKQSAYDSKADIWSLGITAIELAKGEPPHSELHPMKVLFLIPKNNPPTLEGNY 243 Query: 1302 PNNLSDNLKSFIDSCLQSEPARRLTSKQLMNHPFL 1406 S LK F+++CL EP+ R T+K+L+ H F+ Sbjct: 244 ----SKPLKEFVEACLNKEPSFRPTAKELLKHKFI 274
>sp|Q99JT2|MST4_MOUSE Serine/threonine-protein kinase MST4 (STE20-like kinase MST4) (MST-4) (Mammalian STE20-like protein kinase 4) Length = 416 Score = 177 bits (448), Expect = 9e-44 Identities = 102/277 (36%), Positives = 161/277 (58%), Gaps = 3/277 (1%) Frame = +3 Query: 591 NVEPDFENLVKLEKLGKGSFGTVFKCVSIETKGEMAVKKLKIPTTDKTKEEKLSVERELQ 770 N+ E KLE++GKGSFG VFK + T+ +A+K + + ++ ++E +++E+ Sbjct: 16 NIADPEELFTKLERIGKGSFGEVFKGIDNRTQQVVAIKIIDL---EEAEDEIEDIQQEIT 72 Query: 771 VYSKHSDHPRIVKYERNFKTEDAIFLVMEYMKGGSLGGYIRKNGPLSENKIKKYSKQILE 950 V S+ D + KY ++ ++++MEY+ GGS +R GP E +I K+IL+ Sbjct: 73 VLSQ-CDSSYVTKYYGSYLKGSKLWIIMEYLGGGSALDLLRA-GPFDEFQIATMLKEILK 130 Query: 951 ALQYLHEQKILHRDIKSDNILLDEDDNVKLCDFGLSKILTDLAAKDFLNGAIQSSVGTPN 1130 L YLH +K +HRDIK+ N+LL E +VKL DFG++ LTD K + VGTP Sbjct: 131 GLDYLHSEKKIHRDIKAANVLLSEQGDVKLADFGVAGQLTDTQIKR------NTFVGTPF 184 Query: 1131 YMAPEIMNEEPYGSSADVWSFGATMVEMVTGFPPYHQMGVCKVMKIISESRKIEYNYPNN 1310 +MAPE++ + Y S AD+WS G T +E+ G PP M +V+ +I ++ N P Sbjct: 185 WMAPEVIQQSAYDSKADIWSLGITAIELAKGEPPNSDMHPMRVLFLIPKN-----NPPTL 239 Query: 1311 LSD---NLKSFIDSCLQSEPARRLTSKQLMNHPFLRK 1412 + D + K FID+CL +P+ R T+K+L+ H F+ K Sbjct: 240 IGDFTKSFKEFIDACLNKDPSFRPTAKELLKHKFIVK 276
>sp|Q9P289|MST4_HUMAN Serine/threonine-protein kinase MST4 (STE20-like kinase MST4) (MST-4) (Mammalian STE20-like protein kinase 4) (Serine/threonine-protein kinase MASK) (Mst3 and SOK1-related kinase) Length = 416 Score = 176 bits (447), Expect = 1e-43 Identities = 102/277 (36%), Positives = 161/277 (58%), Gaps = 3/277 (1%) Frame = +3 Query: 591 NVEPDFENLVKLEKLGKGSFGTVFKCVSIETKGEMAVKKLKIPTTDKTKEEKLSVERELQ 770 N+ E KLE++GKGSFG VFK + T+ +A+K + + ++ ++E +++E+ Sbjct: 16 NIADPEELFTKLERIGKGSFGEVFKGIDNRTQQVVAIKIIDL---EEAEDEIEDIQQEIT 72 Query: 771 VYSKHSDHPRIVKYERNFKTEDAIFLVMEYMKGGSLGGYIRKNGPLSENKIKKYSKQILE 950 V S+ D + KY ++ ++++MEY+ GGS +R GP E +I K+IL+ Sbjct: 73 VLSQ-CDSSYVTKYYGSYLKGSKLWIIMEYLGGGSALDLLRA-GPFDEFQIATMLKEILK 130 Query: 951 ALQYLHEQKILHRDIKSDNILLDEDDNVKLCDFGLSKILTDLAAKDFLNGAIQSSVGTPN 1130 L YLH +K +HRDIK+ N+LL E +VKL DFG++ LTD K + VGTP Sbjct: 131 GLDYLHSEKKIHRDIKAANVLLSEQGDVKLADFGVAGQLTDTQIKR------NTFVGTPF 184 Query: 1131 YMAPEIMNEEPYGSSADVWSFGATMVEMVTGFPPYHQMGVCKVMKIISESRKIEYNYPNN 1310 +MAPE++ + Y S AD+WS G T +E+ G PP M +V+ +I ++ N P Sbjct: 185 WMAPEVIQQSAYDSKADIWSLGITAIELAKGEPPNSDMHPMRVLFLIPKN-----NPPTL 239 Query: 1311 LSD---NLKSFIDSCLQSEPARRLTSKQLMNHPFLRK 1412 + D + K FID+CL +P+ R T+K+L+ H F+ K Sbjct: 240 VGDFTKSFKEFIDACLNKDPSFRPTAKELLKHKFIVK 276
>sp|O22040|M3K1_ARATH Mitogen-activated protein kinase kinase kinase 1 (Arabidospsis NPK1-related protein kinase 1) Length = 666 Score = 175 bits (444), Expect = 3e-43 Identities = 96/265 (36%), Positives = 157/265 (59%), Gaps = 3/265 (1%) Frame = +3 Query: 621 KLEKLGKGSFGTVFKCVSIETKGEMAVKKLKIPTTDKTKEEKLSVERELQV---YSKHSD 791 K + +G+G+FGTV+ +++++ +AVK++ I +KE+ + +EL+ K+ Sbjct: 71 KGQLIGRGAFGTVYMGMNLDSGELLAVKQVLIAANFASKEKTQAHIQELEEEVKLLKNLS 130 Query: 792 HPRIVKYERNFKTEDAIFLVMEYMKGGSLGGYIRKNGPLSENKIKKYSKQILEALQYLHE 971 HP IV+Y + +D + +++E++ GGS+ + K GP E+ ++ Y++Q+L L+YLH Sbjct: 131 HPNIVRYLGTVREDDTLNILLEFVPGGSISSLLEKFGPFPESVVRTYTRQLLLGLEYLHN 190 Query: 972 QKILHRDIKSDNILLDEDDNVKLCDFGLSKILTDLAAKDFLNGAIQSSVGTPNYMAPEIM 1151 I+HRDIK NIL+D +KL DFG SK + +LA + GA +S GTP +MAPE++ Sbjct: 191 HAIMHRDIKGANILVDNKGCIKLADFGASKQVAELAT---MTGA-KSMKGTPYWMAPEVI 246 Query: 1152 NEEPYGSSADVWSFGATMVEMVTGFPPYHQMGVCKVMKIISESRKIEYNYPNNLSDNLKS 1331 + + SAD+WS G T++EMVTG P+ Q + K P+ LS + K Sbjct: 247 LQTGHSFSADIWSVGCTVIEMVTGKAPWSQQYKEVAAIFFIGTTKSHPPIPDTLSSDAKD 306 Query: 1332 FIDSCLQSEPARRLTSKQLMNHPFL 1406 F+ CLQ P R T+ +L+ HPF+ Sbjct: 307 FLLKCLQEVPNLRPTASELLKHPFV 331
>sp|Q9VXE5|PAKM_DROME Serine/threonine-protein kinase PAK mbt (p21-activated kinase related protein) (Mushroom bodies tiny protein) Length = 639 Score = 173 bits (439), Expect = 1e-42 Identities = 101/279 (36%), Positives = 155/279 (55%) Frame = +3 Query: 576 LQLYFNVEPDFENLVKLEKLGKGSFGTVFKCVSIETKGEMAVKKLKIPTTDKTKEEKLSV 755 LQ+ + ENL K+G+GS GTV T ++AVKK+ D K+++ + Sbjct: 355 LQMVVSAGDpreNLDHFNKIGEGSTGTVCIATDKSTGRQVAVKKM-----DLRKQQRREL 409 Query: 756 ERELQVYSKHSDHPRIVKYERNFKTEDAIFLVMEYMKGGSLGGYIRKNGPLSENKIKKYS 935 V + HP IV+ +F D +++VMEY++GG+L + + + E +I Sbjct: 410 LFNEVVIMRDYHHPNIVETYSSFLVNDELWVVMEYLEGGALTDIVT-HSRMDEEQIATVC 468 Query: 936 KQILEALQYLHEQKILHRDIKSDNILLDEDDNVKLCDFGLSKILTDLAAKDFLNGAIQSS 1115 KQ L+AL YLH Q ++HRDIKSD+ILL D VKL DFG ++ K +S Sbjct: 469 KQCLKALAYLHSQGVIHRDIKSDSILLAADGRVKLSDFGFCAQVSQELPKR------KSL 522 Query: 1116 VGTPNYMAPEIMNEEPYGSSADVWSFGATMVEMVTGFPPYHQMGVCKVMKIISESRKIEY 1295 VGTP +M+PE+++ PYG D+WS G ++EMV G PP+ + M+ I + + Sbjct: 523 VGTPYWMSPEVISRLPYGPEVDIWSLGIMVIEMVDGEPPFFNEPPLQAMRRIRDMQPPNL 582 Query: 1296 NYPNNLSDNLKSFIDSCLQSEPARRLTSKQLMNHPFLRK 1412 + +S L+SF+D L +PA+R T+ +L+ HPFLR+ Sbjct: 583 KNAHKVSPRLQSFLDRMLVRDPAQRATAAELLAHPFLRQ 621
>sp|O22042|M3K3_ARATH Mitogen-activated protein kinase kinase kinase 3 (Arabidospsis NPK1-related protein kinase 3) Length = 651 Score = 173 bits (438), Expect = 1e-42 Identities = 97/265 (36%), Positives = 154/265 (58%), Gaps = 3/265 (1%) Frame = +3 Query: 621 KLEKLGKGSFGTVFKCVSIETKGEMAVKKLKIPTTDKTKEEKLSVERELQV---YSKHSD 791 K E +G G+FG V+ +++++ +A+K++ I + +KE+ REL+ K+ Sbjct: 70 KGELIGCGAFGRVYMGMNLDSGELLAIKQVLIAPSSASKEKTQGHIRELEEEVQLLKNLS 129 Query: 792 HPRIVKYERNFKTEDAIFLVMEYMKGGSLGGYIRKNGPLSENKIKKYSKQILEALQYLHE 971 HP IV+Y + D++ ++ME++ GGS+ + K G E I Y+KQ+L L+YLH Sbjct: 130 HPNIVRYLGTVRESDSLNILMEFVPGGSISSLLEKFGSFPEPVIIMYTKQLLLGLEYLHN 189 Query: 972 QKILHRDIKSDNILLDEDDNVKLCDFGLSKILTDLAAKDFLNGAIQSSVGTPNYMAPEIM 1151 I+HRDIK NIL+D ++L DFG SK + +LA +NGA +S GTP +MAPE++ Sbjct: 190 NGIMHRDIKGANILVDNKGCIRLADFGASKKVVELAT---VNGA-KSMKGTPYWMAPEVI 245 Query: 1152 NEEPYGSSADVWSFGATMVEMVTGFPPYHQMGVCKVMKIISESRKIEYNYPNNLSDNLKS 1331 + + SAD+WS G T++EM TG PP+ + + K P +LS K Sbjct: 246 LQTGHSFSADIWSVGCTVIEMATGKPPWSEQYQQFAAVLHIGRTKAHPPIPEDLSPEAKD 305 Query: 1332 FIDSCLQSEPARRLTSKQLMNHPFL 1406 F+ CL EP+ RL++ +L+ HPF+ Sbjct: 306 FLMKCLHKEPSLRLSATELLQHPFV 330
>sp|Q9FZ36|M3K2_ARATH Mitogen-activated protein kinase kinase kinase 2 (Arabidospsis NPK1-related protein kinase 2) Length = 651 Score = 172 bits (436), Expect = 2e-42 Identities = 95/267 (35%), Positives = 161/267 (60%), Gaps = 5/267 (1%) Frame = +3 Query: 621 KLEKLGKGSFGTVFKCVSIETKGEMAVKKLKIPTTDKTKEEKLSVERELQV---YSKHSD 791 K + +G+G+FGTV+ +++++ +AVK++ I + +KE+ + +EL+ K+ Sbjct: 70 KGQLIGRGAFGTVYMGMNLDSGELLAVKQVLITSNCASKEKTQAHIQELEEEVKLLKNLS 129 Query: 792 HPRIVKYERNFKTEDAIFLVMEYMKGGSLGGYIRKNGPLSENKIKKYSKQILEALQYLHE 971 HP IV+Y + ++ + +++E++ GGS+ + K G E+ ++ Y+ Q+L L+YLH Sbjct: 130 HPNIVRYLGTVREDETLNILLEFVPGGSISSLLEKFGAFPESVVRTYTNQLLLGLEYLHN 189 Query: 972 QKILHRDIKSDNILLDEDDNVKLCDFGLSKILTDLAAKDFLNGAIQSSVGTPNYMAPEIM 1151 I+HRDIK NIL+D +KL DFG SK + +LA ++GA +S GTP +MAPE++ Sbjct: 190 HAIMHRDIKGANILVDNQGCIKLADFGASKQVAELAT---ISGA-KSMKGTPYWMAPEVI 245 Query: 1152 NEEPYGSSADVWSFGATMVEMVTGFPPYHQM--GVCKVMKIISESRKIEYNYPNNLSDNL 1325 + + SAD+WS G T++EMVTG P+ Q + + I + K P+N+S + Sbjct: 246 LQTGHSFSADIWSVGCTVIEMVTGKAPWSQQYKEIAAIFHI--GTTKSHPPIPDNISSDA 303 Query: 1326 KSFIDSCLQSEPARRLTSKQLMNHPFL 1406 F+ CLQ EP R T+ +L+ HPF+ Sbjct: 304 NDFLLKCLQQEPNLRPTASELLKHPFV 330
>sp|Q13177|PAK2_HUMAN Serine/threonine-protein kinase PAK 2 (p21-activated kinase 2) (PAK-2) (PAK65) (Gamma-PAK) (S6/H4 kinase) Length = 524 Score = 172 bits (436), Expect = 2e-42 Identities = 97/305 (31%), Positives = 167/305 (54%) Frame = +3 Query: 495 LDKNRSKPIESDNDIPKPYCLQSRTDDLQLYFNVEPDFENLVKLEKLGKGSFGTVFKCVS 674 LDK + KP +D +I + L+ ++ + + EK+G+G+ GTVF Sbjct: 218 LDKQKKKPKMTDEEI---------MEKLRTIVSIGDPKKKYTRYEKIGQGASGTVFTATD 268 Query: 675 IETKGEMAVKKLKIPTTDKTKEEKLSVERELQVYSKHSDHPRIVKYERNFKTEDAIFLVM 854 + E+A+K++ + K +++L + L + K +P IV + ++ D +F+VM Sbjct: 269 VALGQEVAIKQINL---QKQPKKELIINEILVM--KELKNPNIVNFLDSYLVGDELFVVM 323 Query: 855 EYMKGGSLGGYIRKNGPLSENKIKKYSKQILEALQYLHEQKILHRDIKSDNILLDEDDNV 1034 EY+ GGSL + + + E +I ++ L+AL++LH +++HRDIKSDN+LL + +V Sbjct: 324 EYLAGGSLTDVVTETC-MDEAQIAAVCRECLQALEFLHANQVIHRDIKSDNVLLGMEGSV 382 Query: 1035 KLCDFGLSKILTDLAAKDFLNGAIQSSVGTPNYMAPEIMNEEPYGSSADVWSFGATMVEM 1214 KL DFG +T +K + VGTP +MAPE++ + YG D+WS G +EM Sbjct: 383 KLTDFGFCAQITPEQSKR------STMVGTPYWMAPEVVTRKAYGPKVDIWSLGIMAIEM 436 Query: 1215 VTGFPPYHQMGVCKVMKIISESRKIEYNYPNNLSDNLKSFIDSCLQSEPARRLTSKQLMN 1394 V G PPY + + +I+ + E P LS + F++ CL+ + +R ++K+L+ Sbjct: 437 VEGEPPYLNENPLRALYLIATNGTPELQNPEKLSPIFRDFLNRCLEMDVEKRGSAKELLQ 496 Query: 1395 HPFLR 1409 HPFL+ Sbjct: 497 HPFLK 501
>sp|Q13043|STK4_HUMAN Serine/threonine-protein kinase 4 (STE20-like kinase MST1) (MST-1) (Mammalian STE20-like protein kinase 1) (Serine/threonine-protein kinase Krs-2) Length = 487 Score = 171 bits (434), Expect = 4e-42 Identities = 96/270 (35%), Positives = 155/270 (57%), Gaps = 1/270 (0%) Frame = +3 Query: 624 LEKLGKGSFGTVFKCVSIETKGEMAVKKLKIPTTDKTKEEKLSVERELQVYSKHSDHPRI 803 LEKLG+GS+G+V+K + ET +A+K++ + + + +++S+ ++ D P + Sbjct: 33 LEKLGEGSYGSVYKAIHKETGQIVAIKQVPVESDLQEIIKEISIMQQC-------DSPHV 85 Query: 804 VKYERNFKTEDAIFLVMEYMKGGSLGGYIR-KNGPLSENKIKKYSKQILEALQYLHEQKI 980 VKY ++ +++VMEY GS+ IR +N L+E++I + L+ L+YLH + Sbjct: 86 VKYYGSYFKNTDLWIVMEYCGAGSVSDIIRLRNKTLTEDEIATILQSTLKGLEYLHFMRK 145 Query: 981 LHRDIKSDNILLDEDDNVKLCDFGLSKILTDLAAKDFLNGAIQSSVGTPNYMAPEIMNEE 1160 +HRDIK+ NILL+ + + KL DFG++ LTD AK + +GTP +MAPE++ E Sbjct: 146 IHRDIKAGNILLNTEGHAKLADFGVAGQLTDTMAKR------NTVIGTPFWMAPEVIQEI 199 Query: 1161 PYGSSADVWSFGATMVEMVTGFPPYHQMGVCKVMKIISESRKIEYNYPNNLSDNLKSFID 1340 Y AD+WS G T +EM G PPY + + + +I + + P SDN F+ Sbjct: 200 GYNCVADIWSLGITAIEMAEGKPPYADIHPMRAIFMIPTNPPPTFRKPELWSDNFTDFVK 259 Query: 1341 SCLQSEPARRLTSKQLMNHPFLRKFELVFI 1430 CL P +R T+ QL+ HPF+R + V I Sbjct: 260 QCLVKSPEQRATATQLLQHPFVRSAKGVSI 289
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 160,892,461
Number of Sequences: 369166
Number of extensions: 3465852
Number of successful extensions: 14737
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 10197
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 11035
length of database: 68,354,980
effective HSP length: 115
effective length of database: 47,110,455
effective search space used: 17901972900
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail