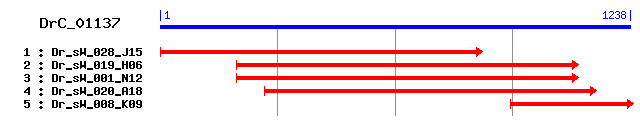
DrC_01137
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_01137 (1238 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q16956|GRP78_APLCA 78 kDa glucose-regulated protein prec... 562 e-160 sp|P06761|GRP78_RAT 78 kDa glucose-regulated protein precur... 558 e-158 sp|P11021|GRP78_HUMAN 78 kDa glucose-regulated protein prec... 556 e-158 sp|Q90593|GRP78_CHICK 78 kDa glucose-regulated protein prec... 556 e-158 sp|P07823|GRP78_MESAU 78 kDa glucose-regulated protein prec... 555 e-157 sp|P20029|GRP78_MOUSE 78 kDa glucose-regulated protein prec... 555 e-157 sp|Q91883|GRP78_XENLA 78 kDa glucose-regulated protein prec... 550 e-156 sp|P19208|HSP7C_CAEBR Heat shock 70 kDa protein C precursor 549 e-156 sp|P27420|HSP7C_CAEEL Heat shock 70 kDa protein C precursor 548 e-155 sp|P29844|HSP7C_DROME Heat shock 70 kDa protein cognate 3 p... 532 e-151
>sp|Q16956|GRP78_APLCA 78 kDa glucose-regulated protein precursor (GRP 78) (BiP) (Protein 1603) Length = 667 Score = 562 bits (1448), Expect = e-160 Identities = 283/367 (77%), Positives = 318/367 (86%) Frame = +3 Query: 3 KKGKDISDDKRAVQKLRREVEKAKRALSNAHSARIEIESLYDGEDFSETLTRARFEELNM 182 KKGKDI D RAVQKLRREVEKAKRALS+AH R+EIES +DGEDFSE+LTRA+FEELNM Sbjct: 282 KKGKDIRKDNRAVQKLRREVEKAKRALSSAHQVRLEIESFFDGEDFSESLTRAKFEELNM 341 Query: 183 DLFRSTMKPVEKVLQDAEMKKEDINEIVLVGGSTRIPKVQQLVKEFFNGKEPSRGINPDE 362 DLFRSTMKPV++VL+DA++K +DI+EIVLVGGSTRIPKVQQLVKE+FNGKEPSRGINPDE Sbjct: 342 DLFRSTMKPVKQVLEDADLKTDDIDEIVLVGGSTRIPKVQQLVKEYFNGKEPSRGINPDE 401 Query: 363 XXXXXXXXXXXXXXXXEETGDIVLLDVCPLTLGIETVGGVMTKLIPRNTVIPTKKSQVFS 542 E+TGD+VLLDV PLT+GIETVGGVMTKLIPRNTVIPTKKSQ+FS Sbjct: 402 AVAYGAAVQAGVLSGEEDTGDLVLLDVNPLTMGIETVGGVMTKLIPRNTVIPTKKSQIFS 461 Query: 543 TAADNQPTVTIQVFEGERPMTKDNHFLGKFDLTGIPAAPRGVPQIEVTFEIDVNGILKVS 722 TAADNQPTVTIQV+EGER MTKDNH LGKFDLTGIP APRGVPQIEVTFEIDVNGILKV+ Sbjct: 462 TAADNQPTVTIQVYEGERSMTKDNHLLGKFDLTGIPPAPRGVPQIEVTFEIDVNGILKVT 521 Query: 723 AEDKGTGNKNKIVIQKDQNRLNTEDIEKMIKDAETYADQDKKVKERVEAKNDLEGYAYSL 902 AEDKGTG+KN+IVIQ DQNRL+ EDIE+MI DAE YAD+DKKVKE+V+AKN+LE YAYSL Sbjct: 522 AEDKGTGSKNQIVIQNDQNRLSPEDIERMINDAEKYADEDKKVKEKVDAKNELESYAYSL 581 Query: 903 KTQINDKEKLGGKLSPSDKETITKATEEAISWLEKNQQADSEELKAQKKKVEDIVTPIMT 1082 K QI DKEKLG KLS DKE IT+A +EAI WLE N +A+SE +K ++E IV PIMT Sbjct: 582 KNQIGDKEKLGAKLSDEDKEKITEAVDEAIKWLESNAEAESEAFNEKKTELEGIVQPIMT 641 Query: 1083 KLYQQSG 1103 KLY+QSG Sbjct: 642 KLYEQSG 648
>sp|P06761|GRP78_RAT 78 kDa glucose-regulated protein precursor (GRP 78) (Immunoglobulin heavy chain binding protein) (BiP) (Steroidogenesis-activator polypeptide) Length = 654 Score = 558 bits (1437), Expect = e-158 Identities = 273/367 (74%), Positives = 316/367 (86%) Frame = +3 Query: 3 KKGKDISDDKRAVQKLRREVEKAKRALSNAHSARIEIESLYDGEDFSETLTRARFEELNM 182 K GKD+ D RAVQKLRREVEKAKRALS+ H ARIEIES ++GEDFSETLTRA+FEELNM Sbjct: 273 KTGKDVRKDNRAVQKLRREVEKAKRALSSQHQARIEIESFFEGEDFSETLTRAKFEELNM 332 Query: 183 DLFRSTMKPVEKVLQDAEMKKEDINEIVLVGGSTRIPKVQQLVKEFFNGKEPSRGINPDE 362 DLFRSTMKPV+KVL+D+++KK DI+EIVLVGGSTRIPK+QQLVKEFFNGKEPSRGINPDE Sbjct: 333 DLFRSTMKPVQKVLEDSDLKKSDIDEIVLVGGSTRIPKIQQLVKEFFNGKEPSRGINPDE 392 Query: 363 XXXXXXXXXXXXXXXXEETGDIVLLDVCPLTLGIETVGGVMTKLIPRNTVIPTKKSQVFS 542 ++TGD+VLLDVCPLTLGIETVGGVMTKLIPRNTV+PTKKSQ+FS Sbjct: 393 AVAYGAAVQAGVLSGDQDTGDLVLLDVCPLTLGIETVGGVMTKLIPRNTVVPTKKSQIFS 452 Query: 543 TAADNQPTVTIQVFEGERPMTKDNHFLGKFDLTGIPAAPRGVPQIEVTFEIDVNGILKVS 722 TA+DNQPTVTI+V+EGERP+TKDNH LG FDLTGIP APRGVPQIEVTFEIDVNGIL+V+ Sbjct: 453 TASDNQPTVTIKVYEGERPLTKDNHLLGTFDLTGIPPAPRGVPQIEVTFEIDVNGILRVT 512 Query: 723 AEDKGTGNKNKIVIQKDQNRLNTEDIEKMIKDAETYADQDKKVKERVEAKNDLEGYAYSL 902 AEDKGTGNKNKI I DQNRL E+IE+M+ DAE +A++DKK+KER++ +N+LE YAYSL Sbjct: 513 AEDKGTGNKNKITITNDQNRLTPEEIERMVNDAEKFAEEDKKLKERIDTRNELESYAYSL 572 Query: 903 KTQINDKEKLGGKLSPSDKETITKATEEAISWLEKNQQADSEELKAQKKKVEDIVTPIMT 1082 K QI DKEKLGGKLSP DKET+ KA EE I WLE +Q AD E+ KA+KK++E+IV PI++ Sbjct: 573 KNQIGDKEKLGGKLSPEDKETMEKAVEEKIEWLESHQDADIEDFKAKKKELEEIVQPIIS 632 Query: 1083 KLYQQSG 1103 KLY G Sbjct: 633 KLYGSGG 639
>sp|P11021|GRP78_HUMAN 78 kDa glucose-regulated protein precursor (GRP 78) (Immunoglobulin heavy chain binding protein) (BiP) (Endoplasmic reticulum lumenal Ca(2+) binding protein grp78) Length = 654 Score = 556 bits (1434), Expect = e-158 Identities = 273/367 (74%), Positives = 316/367 (86%) Frame = +3 Query: 3 KKGKDISDDKRAVQKLRREVEKAKRALSNAHSARIEIESLYDGEDFSETLTRARFEELNM 182 K GKD+ D RAVQKLRREVEKAKRALS+ H ARIEIES Y+GEDFSETLTRA+FEELNM Sbjct: 273 KTGKDVRKDNRAVQKLRREVEKAKRALSSQHQARIEIESFYEGEDFSETLTRAKFEELNM 332 Query: 183 DLFRSTMKPVEKVLQDAEMKKEDINEIVLVGGSTRIPKVQQLVKEFFNGKEPSRGINPDE 362 DLFRSTMKPV+KVL+D+++KK DI+EIVLVGGSTRIPK+QQLVKEFFNGKEPSRGINPDE Sbjct: 333 DLFRSTMKPVQKVLEDSDLKKSDIDEIVLVGGSTRIPKIQQLVKEFFNGKEPSRGINPDE 392 Query: 363 XXXXXXXXXXXXXXXXEETGDIVLLDVCPLTLGIETVGGVMTKLIPRNTVIPTKKSQVFS 542 ++TGD+VLLDVCPLTLGIETVGGVMTKLIPRNTV+PTKKSQ+FS Sbjct: 393 AVAYGAAVQAGVLSGDQDTGDLVLLDVCPLTLGIETVGGVMTKLIPRNTVVPTKKSQIFS 452 Query: 543 TAADNQPTVTIQVFEGERPMTKDNHFLGKFDLTGIPAAPRGVPQIEVTFEIDVNGILKVS 722 TA+DNQPTVTI+V+EGERP+TKDNH LG FDLTGIP APRGVPQIEVTFEIDVNGIL+V+ Sbjct: 453 TASDNQPTVTIKVYEGERPLTKDNHLLGTFDLTGIPPAPRGVPQIEVTFEIDVNGILRVT 512 Query: 723 AEDKGTGNKNKIVIQKDQNRLNTEDIEKMIKDAETYADQDKKVKERVEAKNDLEGYAYSL 902 AEDKGTGNKNKI I DQNRL E+IE+M+ DAE +A++DKK+KER++ +N+LE YAYSL Sbjct: 513 AEDKGTGNKNKITITNDQNRLTPEEIERMVNDAEKFAEEDKKLKERIDTRNELESYAYSL 572 Query: 903 KTQINDKEKLGGKLSPSDKETITKATEEAISWLEKNQQADSEELKAQKKKVEDIVTPIMT 1082 K QI DKEKLGGKLS DKET+ KA EE I WLE +Q AD E+ KA+KK++E+IV PI++ Sbjct: 573 KNQIGDKEKLGGKLSSEDKETMEKAVEEKIEWLESHQDADIEDFKAKKKELEEIVQPIIS 632 Query: 1083 KLYQQSG 1103 KLY +G Sbjct: 633 KLYGSAG 639
>sp|Q90593|GRP78_CHICK 78 kDa glucose-regulated protein precursor (GRP 78) (Immunoglobulin heavy chain binding protein) (BiP) Length = 652 Score = 556 bits (1433), Expect = e-158 Identities = 272/367 (74%), Positives = 317/367 (86%) Frame = +3 Query: 3 KKGKDISDDKRAVQKLRREVEKAKRALSNAHSARIEIESLYDGEDFSETLTRARFEELNM 182 K GKD+ D RAVQKLRREVEKAKRALS+ H ARIEIES ++GEDFSETLTRA+FEELNM Sbjct: 271 KTGKDVRKDNRAVQKLRREVEKAKRALSSQHQARIEIESFFEGEDFSETLTRAKFEELNM 330 Query: 183 DLFRSTMKPVEKVLQDAEMKKEDINEIVLVGGSTRIPKVQQLVKEFFNGKEPSRGINPDE 362 DLFRSTMKPV+KVL+D+++KK DI+EIVLVGGSTRIPK+QQLVKEFFNGKEPSRGINPDE Sbjct: 331 DLFRSTMKPVQKVLEDSDLKKSDIDEIVLVGGSTRIPKIQQLVKEFFNGKEPSRGINPDE 390 Query: 363 XXXXXXXXXXXXXXXXEETGDIVLLDVCPLTLGIETVGGVMTKLIPRNTVIPTKKSQVFS 542 ++TGD+VLLDVCPLTLGIETVGGVMTKLIPRNTV+PTKKSQ+FS Sbjct: 391 AVAYGAAVQAGVLSGDQDTGDLVLLDVCPLTLGIETVGGVMTKLIPRNTVVPTKKSQIFS 450 Query: 543 TAADNQPTVTIQVFEGERPMTKDNHFLGKFDLTGIPAAPRGVPQIEVTFEIDVNGILKVS 722 TA+DNQPTVTI+V+EGERP+TKDNH LG FDLTGIP APRGVPQIEVTFEIDVNGIL+V+ Sbjct: 451 TASDNQPTVTIKVYEGERPLTKDNHLLGTFDLTGIPPAPRGVPQIEVTFEIDVNGILRVT 510 Query: 723 AEDKGTGNKNKIVIQKDQNRLNTEDIEKMIKDAETYADQDKKVKERVEAKNDLEGYAYSL 902 AEDKGTGNKNKI I DQNRL E+IE+M+ DAE +A++DKK+KER++A+N+LE YAYSL Sbjct: 511 AEDKGTGNKNKITITNDQNRLTPEEIERMVNDAEKFAEEDKKLKERIDARNELESYAYSL 570 Query: 903 KTQINDKEKLGGKLSPSDKETITKATEEAISWLEKNQQADSEELKAQKKKVEDIVTPIMT 1082 K QI DKEKLGGKLS DKETI KA EE I WLE +Q AD E+ K++KK++E++V PI++ Sbjct: 571 KNQIGDKEKLGGKLSSEDKETIEKAVEEKIEWLESHQDADIEDFKSKKKELEEVVQPIVS 630 Query: 1083 KLYQQSG 1103 KLY +G Sbjct: 631 KLYGSAG 637
>sp|P07823|GRP78_MESAU 78 kDa glucose-regulated protein precursor (GRP 78) (Immunoglobulin heavy chain binding protein) (BiP) Length = 654 Score = 555 bits (1430), Expect = e-157 Identities = 272/367 (74%), Positives = 316/367 (86%) Frame = +3 Query: 3 KKGKDISDDKRAVQKLRREVEKAKRALSNAHSARIEIESLYDGEDFSETLTRARFEELNM 182 K GKD+ D RAVQKLRREVEKAKRALS+ H ARIEIES ++GEDFSETLTRA+FEELNM Sbjct: 273 KTGKDVRKDNRAVQKLRREVEKAKRALSSQHQARIEIESFFEGEDFSETLTRAKFEELNM 332 Query: 183 DLFRSTMKPVEKVLQDAEMKKEDINEIVLVGGSTRIPKVQQLVKEFFNGKEPSRGINPDE 362 DLFRSTMKPV+KVL+D+++KK DI+EIVLVGGSTRIPK+QQLVKEFFNGKEPSRGINPDE Sbjct: 333 DLFRSTMKPVQKVLEDSDLKKSDIDEIVLVGGSTRIPKIQQLVKEFFNGKEPSRGINPDE 392 Query: 363 XXXXXXXXXXXXXXXXEETGDIVLLDVCPLTLGIETVGGVMTKLIPRNTVIPTKKSQVFS 542 ++TGD+VLLDVCPLTLGIETVGGVMTKLIPRNTV+PTKKSQ+FS Sbjct: 393 AVAYGAAVQAGVLSGDQDTGDLVLLDVCPLTLGIETVGGVMTKLIPRNTVVPTKKSQIFS 452 Query: 543 TAADNQPTVTIQVFEGERPMTKDNHFLGKFDLTGIPAAPRGVPQIEVTFEIDVNGILKVS 722 TA+DNQPTVTI+V+EGERP+TKDNH LG FDLTGIP APRGVPQIEVTFEIDVNGIL+V+ Sbjct: 453 TASDNQPTVTIKVYEGERPLTKDNHLLGTFDLTGIPPAPRGVPQIEVTFEIDVNGILRVT 512 Query: 723 AEDKGTGNKNKIVIQKDQNRLNTEDIEKMIKDAETYADQDKKVKERVEAKNDLEGYAYSL 902 AEDKGTGNKNKI I DQNRL E+IE+M+ DAE +A++DKK+KER++ +N+LE YAYSL Sbjct: 513 AEDKGTGNKNKITITNDQNRLTPEEIERMVNDAEKFAEEDKKLKERIDTRNELESYAYSL 572 Query: 903 KTQINDKEKLGGKLSPSDKETITKATEEAISWLEKNQQADSEELKAQKKKVEDIVTPIMT 1082 K QI DKEKLGGKLS DKET+ KA EE I WLE +Q AD E+ KA+KK++E+IV PI++ Sbjct: 573 KNQIGDKEKLGGKLSSEDKETMEKAVEEKIEWLESHQDADIEDFKAKKKELEEIVQPIIS 632 Query: 1083 KLYQQSG 1103 KLY +G Sbjct: 633 KLYGSAG 639
>sp|P20029|GRP78_MOUSE 78 kDa glucose-regulated protein precursor (GRP 78) (Immunoglobulin heavy chain binding protein) (BiP) Length = 655 Score = 555 bits (1429), Expect = e-157 Identities = 272/367 (74%), Positives = 315/367 (85%) Frame = +3 Query: 3 KKGKDISDDKRAVQKLRREVEKAKRALSNAHSARIEIESLYDGEDFSETLTRARFEELNM 182 K GKD+ D RAVQKLRREVEKAKRALS+ H ARIEIES ++GEDFSETLTRA+FEELNM Sbjct: 274 KTGKDVRKDNRAVQKLRREVEKAKRALSSQHQARIEIESFFEGEDFSETLTRAKFEELNM 333 Query: 183 DLFRSTMKPVEKVLQDAEMKKEDINEIVLVGGSTRIPKVQQLVKEFFNGKEPSRGINPDE 362 DLFRSTMKPV+KVL+D+++KK DI+EIVLVGGSTRIPK+QQLVKEFFNGKEPSRGINPDE Sbjct: 334 DLFRSTMKPVQKVLEDSDLKKSDIDEIVLVGGSTRIPKIQQLVKEFFNGKEPSRGINPDE 393 Query: 363 XXXXXXXXXXXXXXXXEETGDIVLLDVCPLTLGIETVGGVMTKLIPRNTVIPTKKSQVFS 542 ++TGD+VLLDVCPLTLGIETVGGVMTKLIPRNTV+PTKKSQ+FS Sbjct: 394 AVAYGAAVQAGVLSGDQDTGDLVLLDVCPLTLGIETVGGVMTKLIPRNTVVPTKKSQIFS 453 Query: 543 TAADNQPTVTIQVFEGERPMTKDNHFLGKFDLTGIPAAPRGVPQIEVTFEIDVNGILKVS 722 TA+DNQPTVTI+V+EGERP+TKDNH LG FDLTGIP APRGVPQIEVTFEIDVNGIL+V+ Sbjct: 454 TASDNQPTVTIKVYEGERPLTKDNHLLGTFDLTGIPPAPRGVPQIEVTFEIDVNGILRVT 513 Query: 723 AEDKGTGNKNKIVIQKDQNRLNTEDIEKMIKDAETYADQDKKVKERVEAKNDLEGYAYSL 902 AEDKGTGNKNKI I DQNRL E+IE+M+ DAE +A++DKK+KER++ +N+LE YAYSL Sbjct: 514 AEDKGTGNKNKITITNDQNRLTPEEIERMVNDAEKFAEEDKKLKERIDTRNELESYAYSL 573 Query: 903 KTQINDKEKLGGKLSPSDKETITKATEEAISWLEKNQQADSEELKAQKKKVEDIVTPIMT 1082 K QI DKEKLGGKLS DKET+ KA EE I WLE +Q AD E+ KA+KK++E+IV PI++ Sbjct: 574 KNQIGDKEKLGGKLSSEDKETMEKAVEEKIEWLESHQDADIEDFKAKKKELEEIVQPIIS 633 Query: 1083 KLYQQSG 1103 KLY G Sbjct: 634 KLYGSGG 640
>sp|Q91883|GRP78_XENLA 78 kDa glucose-regulated protein precursor (GRP 78) (Immunoglobulin heavy chain binding protein) (BiP) Length = 658 Score = 550 bits (1417), Expect = e-156 Identities = 272/367 (74%), Positives = 313/367 (85%) Frame = +3 Query: 3 KKGKDISDDKRAVQKLRREVEKAKRALSNAHSARIEIESLYDGEDFSETLTRARFEELNM 182 K GKD+ DKRAVQKLRREVEKAKRALS H +RIEIES ++GEDFSETLTRA+FEELNM Sbjct: 274 KTGKDVRADKRAVQKLRREVEKAKRALSAQHQSRIEIESFFEGEDFSETLTRAKFEELNM 333 Query: 183 DLFRSTMKPVEKVLQDAEMKKEDINEIVLVGGSTRIPKVQQLVKEFFNGKEPSRGINPDE 362 DLFRSTMKPV+KVL D+++KK DI+EIVLVGGSTRIPK+QQLVKE FNGKEPSRGINPDE Sbjct: 334 DLFRSTMKPVQKVLDDSDLKKSDIDEIVLVGGSTRIPKIQQLVKELFNGKEPSRGINPDE 393 Query: 363 XXXXXXXXXXXXXXXXEETGDIVLLDVCPLTLGIETVGGVMTKLIPRNTVIPTKKSQVFS 542 ++TGD+VLLDVCPLTLGIETVGGVMTKLIPRNTV+PTKKSQ+FS Sbjct: 394 AVAYGAAVQAGVLSGDQDTGDLVLLDVCPLTLGIETVGGVMTKLIPRNTVVPTKKSQIFS 453 Query: 543 TAADNQPTVTIQVFEGERPMTKDNHFLGKFDLTGIPAAPRGVPQIEVTFEIDVNGILKVS 722 TA+DNQPTVTI+V+EGERP+TKDNH LG FDLTGIP APRGVPQIEVTFEIDVNGIL+V+ Sbjct: 454 TASDNQPTVTIKVYEGERPLTKDNHLLGTFDLTGIPPAPRGVPQIEVTFEIDVNGILRVT 513 Query: 723 AEDKGTGNKNKIVIQKDQNRLNTEDIEKMIKDAETYADQDKKVKERVEAKNDLEGYAYSL 902 AEDKGTGNKNKI I DQNRL E+IE+M+ DAE +A++DKK+KER++ +N+LE YAYSL Sbjct: 514 AEDKGTGNKNKITITNDQNRLTPEEIERMVTDAEKFAEEDKKLKERIDTRNELESYAYSL 573 Query: 903 KTQINDKEKLGGKLSPSDKETITKATEEAISWLEKNQQADSEELKAQKKKVEDIVTPIMT 1082 K QI DKEKLGGKLS DKETI KA EE I WLE +Q AD E+ KA+KK++E+IV PI+ Sbjct: 574 KNQIGDKEKLGGKLSSEDKETIEKAVEEKIEWLESHQDADIEDFKAKKKELEEIVQPIVG 633 Query: 1083 KLYQQSG 1103 KLY +G Sbjct: 634 KLYGGAG 640
>sp|P19208|HSP7C_CAEBR Heat shock 70 kDa protein C precursor Length = 661 Score = 549 bits (1415), Expect = e-156 Identities = 276/367 (75%), Positives = 312/367 (85%) Frame = +3 Query: 3 KKGKDISDDKRAVQKLRREVEKAKRALSNAHSARIEIESLYDGEDFSETLTRARFEELNM 182 K GKD+ DKRAVQKLRREVEKAKRALS H ++EIESL+DGEDFSETLTRA+FEELNM Sbjct: 278 KSGKDLRKDKRAVQKLRREVEKAKRALSTQHQTKVEIESLFDGEDFSETLTRAKFEELNM 337 Query: 183 DLFRSTMKPVEKVLQDAEMKKEDINEIVLVGGSTRIPKVQQLVKEFFNGKEPSRGINPDE 362 DLFR+T+KPV+KVL+D+++KK+D++EIVLVGGSTRIPKVQQL+KEFFNGKEPSRGINPDE Sbjct: 338 DLFRATLKPVQKVLEDSDLKKDDVHEIVLVGGSTRIPKVQQLIKEFFNGKEPSRGINPDE 397 Query: 363 XXXXXXXXXXXXXXXXEETGDIVLLDVCPLTLGIETVGGVMTKLIPRNTVIPTKKSQVFS 542 E+TG+IVLLDV PLT+GIETVGGVMTKLI RNTVIPTKKSQVFS Sbjct: 398 AVAYGAAVQGGVISGEEDTGEIVLLDVNPLTMGIETVGGVMTKLISRNTVIPTKKSQVFS 457 Query: 543 TAADNQPTVTIQVFEGERPMTKDNHFLGKFDLTGIPAAPRGVPQIEVTFEIDVNGILKVS 722 TAADNQPTVTIQVFEGERPMTKDNH LGKFDLTGIP APRGVPQIEVTFEIDVNGIL V+ Sbjct: 458 TAADNQPTVTIQVFEGERPMTKDNHQLGKFDLTGIPPAPRGVPQIEVTFEIDVNGILHVT 517 Query: 723 AEDKGTGNKNKIVIQKDQNRLNTEDIEKMIKDAETYADQDKKVKERVEAKNDLEGYAYSL 902 AEDKGTGNKNKI I DQNRL+ EDIE+MI DAE +A+ DKKVKE+ EA+N+LE YAYSL Sbjct: 518 AEDKGTGNKNKITITNDQNRLSPEDIERMINDAEKFAEDDKKVKEKAEARNELESYAYSL 577 Query: 903 KTQINDKEKLGGKLSPSDKETITKATEEAISWLEKNQQADSEELKAQKKKVEDIVTPIMT 1082 K QI DKEKLGGKL DK+TI +A +EAISWL N A +EELK QKK++E V PI++ Sbjct: 578 KNQIGDKEKLGGKLDEDDKKTIEEAVDEAISWLGSNADASAEELKEQKKELEGKVQPIVS 637 Query: 1083 KLYQQSG 1103 KLY+ G Sbjct: 638 KLYKDGG 644
>sp|P27420|HSP7C_CAEEL Heat shock 70 kDa protein C precursor Length = 661 Score = 548 bits (1411), Expect = e-155 Identities = 274/367 (74%), Positives = 313/367 (85%) Frame = +3 Query: 3 KKGKDISDDKRAVQKLRREVEKAKRALSNAHSARIEIESLYDGEDFSETLTRARFEELNM 182 K GKD+ DKRAVQKLRREVEKAKRALS H ++EIESL+DGEDFSETLTRA+FEELNM Sbjct: 278 KSGKDLRKDKRAVQKLRREVEKAKRALSTQHQTKVEIESLFDGEDFSETLTRAKFEELNM 337 Query: 183 DLFRSTMKPVEKVLQDAEMKKEDINEIVLVGGSTRIPKVQQLVKEFFNGKEPSRGINPDE 362 DLFR+T+KPV+KVL+D+++KK+D++EIVLVGGSTRIPKVQQL+KEFFNGKEPSRGINPDE Sbjct: 338 DLFRATLKPVQKVLEDSDLKKDDVHEIVLVGGSTRIPKVQQLIKEFFNGKEPSRGINPDE 397 Query: 363 XXXXXXXXXXXXXXXXEETGDIVLLDVCPLTLGIETVGGVMTKLIPRNTVIPTKKSQVFS 542 E+TG+IVLLDV PLT+GIETVGGVMTKLI RNTVIPTKKSQVFS Sbjct: 398 AVAYGAAVQGGVISGEEDTGEIVLLDVNPLTMGIETVGGVMTKLIGRNTVIPTKKSQVFS 457 Query: 543 TAADNQPTVTIQVFEGERPMTKDNHFLGKFDLTGIPAAPRGVPQIEVTFEIDVNGILKVS 722 TAADNQPTVTIQVFEGERPMTKDNH LGKFDLTG+P APRGVPQIEVTFEIDVNGIL V+ Sbjct: 458 TAADNQPTVTIQVFEGERPMTKDNHQLGKFDLTGLPPAPRGVPQIEVTFEIDVNGILHVT 517 Query: 723 AEDKGTGNKNKIVIQKDQNRLNTEDIEKMIKDAETYADQDKKVKERVEAKNDLEGYAYSL 902 AEDKGTGNKNKI I DQNRL+ EDIE+MI DAE +A+ DKKVK++ EA+N+LE YAY+L Sbjct: 518 AEDKGTGNKNKITITNDQNRLSPEDIERMINDAEKFAEDDKKVKDKAEARNELESYAYNL 577 Query: 903 KTQINDKEKLGGKLSPSDKETITKATEEAISWLEKNQQADSEELKAQKKKVEDIVTPIMT 1082 K QI DKEKLGGKL DK+TI +A EEAISWL N +A +EELK QKK +E V PI++ Sbjct: 578 KNQIEDKEKLGGKLDEDDKKTIEEAVEEAISWLGSNAEASAEELKEQKKDLESKVQPIVS 637 Query: 1083 KLYQQSG 1103 KLY+ +G Sbjct: 638 KLYKDAG 644
>sp|P29844|HSP7C_DROME Heat shock 70 kDa protein cognate 3 precursor (78 kDa glucose regulated protein homolog) (GRP 78) (Heat shock protein cognate 72) Length = 656 Score = 532 bits (1370), Expect = e-151 Identities = 266/367 (72%), Positives = 307/367 (83%) Frame = +3 Query: 3 KKGKDISDDKRAVQKLRREVEKAKRALSNAHSARIEIESLYDGEDFSETLTRARFEELNM 182 KKGKDI D RAVQKLRREVEKAKRALS +H RIEIES ++G+DFSETLTRA+FEELN+ Sbjct: 273 KKGKDIRKDNRAVQKLRREVEKAKRALSGSHQVRIEIESFFEGDDFSETLTRAKFEELNL 332 Query: 183 DLFRSTMKPVEKVLQDAEMKKEDINEIVLVGGSTRIPKVQQLVKEFFNGKEPSRGINPDE 362 DLFRST+KPV+KVL+DA+M K+D++EIVLVGGSTRIPKVQQLVK+FF GKEPSRGINPDE Sbjct: 333 DLFRSTLKPVQKVLEDADMNKKDVHEIVLVGGSTRIPKVQQLVKDFFGGKEPSRGINPDE 392 Query: 363 XXXXXXXXXXXXXXXXEETGDIVLLDVCPLTLGIETVGGVMTKLIPRNTVIPTKKSQVFS 542 ++T IVLLDV PLT+GIETVGGVMTKLIPRNTVIPTKKSQVFS Sbjct: 393 AVAYGAAVQAGVLSGEQDTDAIVLLDVNPLTMGIETVGGVMTKLIPRNTVIPTKKSQVFS 452 Query: 543 TAADNQPTVTIQVFEGERPMTKDNHFLGKFDLTGIPAAPRGVPQIEVTFEIDVNGILKVS 722 TA+DNQ TVTIQV+EGERPMTKDNH LGKFDLTGIP APRG+PQIEV+FEID NGIL+VS Sbjct: 453 TASDNQHTVTIQVYEGERPMTKDNHLLGKFDLTGIPPAPRGIPQIEVSFEIDANGILQVS 512 Query: 723 AEDKGTGNKNKIVIQKDQNRLNTEDIEKMIKDAETYADQDKKVKERVEAKNDLEGYAYSL 902 AEDKGTGNK KIVI DQNRL EDI++MI+DAE +AD+DKK+KERVE++N+LE YAYSL Sbjct: 513 AEDKGTGNKEKIVITNDQNRLTPEDIDRMIRDAEKFADEDKKLKERVESRNELESYAYSL 572 Query: 903 KTQINDKEKLGGKLSPSDKETITKATEEAISWLEKNQQADSEELKAQKKKVEDIVTPIMT 1082 K QI DK+KLG KLS +K + A +E+I WLE+N AD EE K QKK +E IV P++ Sbjct: 573 KNQIGDKDKLGAKLSDDEKNKLESAIDESIKWLEQNPDADPEEYKKQKKDLEAIVQPVIA 632 Query: 1083 KLYQQSG 1103 KLYQ +G Sbjct: 633 KLYQGAG 639
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 112,323,213
Number of Sequences: 369166
Number of extensions: 2037469
Number of successful extensions: 9569
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 7930
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 8520
length of database: 68,354,980
effective HSP length: 113
effective length of database: 47,479,925
effective search space used: 14196497575
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail