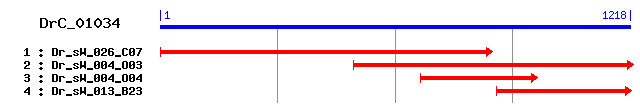
DrC_01034
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_01034 (1218 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|O44451|ODPB_CAEEL Pyruvate dehydrogenase E1 component be... 479 e-135 sp|P11177|ODPB_HUMAN Pyruvate dehydrogenase E1 component be... 475 e-133 sp|Q9D051|ODPB_MOUSE Pyruvate dehydrogenase E1 component be... 469 e-132 sp|P49432|ODPB_RAT Pyruvate dehydrogenase E1 component beta... 468 e-131 sp|P26269|ODPB_ASCSU Pyruvate dehydrogenase E1 component be... 464 e-130 sp|P32473|ODPB_YEAST Pyruvate dehydrogenase E1 component be... 417 e-116 sp|Q38799|ODPB_ARATH Pyruvate dehydrogenase E1 component be... 414 e-115 sp|Q09171|ODPB_SCHPO Pyruvate dehydrogenase E1 component be... 405 e-112 sp|P52904|ODPB_PEA Pyruvate dehydrogenase E1 component beta... 396 e-110 sp|Q9R9N4|ODPB_RHIME Pyruvate dehydrogenase E1 component, b... 390 e-108
>sp|O44451|ODPB_CAEEL Pyruvate dehydrogenase E1 component beta subunit, mitochondrial precursor Length = 352 Score = 479 bits (1234), Expect = e-135 Identities = 232/327 (70%), Positives = 274/327 (83%) Frame = +2 Query: 83 LTVRDALNQALKEEIERDKNVFILGEEVAQYDGAYKVTKGLWKLFGDERVVDTPITEMGF 262 +TVRDALNQA+ EEI+RD VF++GEEVAQYDGAYK++KGLWK GD+RVVDTPITEMGF Sbjct: 25 MTVRDALNQAMDEEIKRDDRVFLMGEEVAQYDGAYKISKGLWKKHGDKRVVDTPITEMGF 84 Query: 263 AGIAVGAAFHGLRPVCEFMTFNFAMQAVDHIINSAAKGYYMSAGQIKVPIVFRGPNGAAA 442 AGIAVGAAF GLRP+CEFMTFNF+MQA+D IINSAAK YYMSAG++ VPIVFRGPNGAAA Sbjct: 85 AGIAVGAAFAGLRPICEFMTFNFSMQAIDQIINSAAKTYYMSAGRVPVPIVFRGPNGAAA 144 Query: 443 GVAAQHSQDFSAWFTHCPALKVLSPYSSEDARGLLKSAIRDPDPVIVLENEVLYGHPFDT 622 GVAAQHSQDFSAW+ HCP LKV+ PYS+EDA+GLLK+AIRD +PV+ LENE+LYG F Sbjct: 145 GVAAQHSQDFSAWYAHCPGLKVVCPYSAEDAKGLLKAAIRDDNPVVFLENEILYGQSFPV 204 Query: 623 PDEALSPDFLIPIGKAKIEKTGTSVTLVSFSKGVDLCLNAANELAKIGVDCEVINLRSLR 802 DE LS DF++PIGKAKIE+ G VT+VS+S+GV+ L AA +L IGV EVINLRSLR Sbjct: 205 GDEVLSDDFVVPIGKAKIERAGDHVTIVSYSRGVEFSLEAAKQLEAIGVSAEVINLRSLR 264 Query: 803 PLDMETVIKSVVKTHHLVTVEESWPTCGIGSEILARVMETQAFNYLDAPALRVTGADIPM 982 P D E++ +SV KTHHLV+VE WP GIGSEI A+VME+ F+ LDAP LRVTG D+PM Sbjct: 265 PFDFESIRQSVHKTHHLVSVETGWPFAGIGSEIAAQVMESDVFDQLDAPLLRVTGVDVPM 324 Query: 983 PYAKPLEMEALPRVENVIKSVKKVLHI 1063 PY + LE ALP E+V+K+VKK L+I Sbjct: 325 PYTQTLEAAALPTAEHVVKAVKKSLNI 351
>sp|P11177|ODPB_HUMAN Pyruvate dehydrogenase E1 component beta subunit, mitochondrial precursor (PDHE1-B) Length = 359 Score = 475 bits (1223), Expect = e-133 Identities = 227/340 (66%), Positives = 288/340 (84%) Frame = +2 Query: 44 QRHIKLSAAINVKLTVRDALNQALKEEIERDKNVFILGEEVAQYDGAYKVTKGLWKLFGD 223 +R +A V++TVRDA+NQ + EE+ERD+ VF+LGEEVAQYDGAYKV++GLWK +GD Sbjct: 20 KRRFHWTAPAAVQVTVRDAINQGMDEELERDEKVFLLGEEVAQYDGAYKVSRGLWKKYGD 79 Query: 224 ERVVDTPITEMGFAGIAVGAAFHGLRPVCEFMTFNFAMQAVDHIINSAAKGYYMSAGQIK 403 +R++DTPI+EMGFAGIAVGAA GLRP+CEFMTFNF+MQA+D +INSAAK YYMS G Sbjct: 80 KRIIDTPISEMGFAGIAVGAAMAGLRPICEFMTFNFSMQAIDQVINSAAKTYYMSGGLQP 139 Query: 404 VPIVFRGPNGAAAGVAAQHSQDFSAWFTHCPALKVLSPYSSEDARGLLKSAIRDPDPVIV 583 VPIVFRGPNGA+AGVAAQHSQ F+AW+ HCP LKV+SP++SEDA+GL+KSAIRD +PV+V Sbjct: 140 VPIVFRGPNGASAGVAAQHSQCFAAWYGHCPGLKVVSPWNSEDAKGLIKSAIRDNNPVVV 199 Query: 584 LENEVLYGHPFDTPDEALSPDFLIPIGKAKIEKTGTSVTLVSFSKGVDLCLNAANELAKI 763 LENE++YG PF+ P EA S DFLIPIGKAKIE+ GT +T+VS S+ V CL AA L+K Sbjct: 200 LENELMYGVPFEFPPEAQSKDFLIPIGKAKIERQGTHITVVSHSRPVGHCLEAAAVLSKE 259 Query: 764 GVDCEVINLRSLRPLDMETVIKSVVKTHHLVTVEESWPTCGIGSEILARVMETQAFNYLD 943 GV+CEVIN+R++RP+DMET+ SV+KT+HLVTVE WP G+G+EI AR+ME AFN+LD Sbjct: 260 GVECEVINMRTIRPMDMETIEASVMKTNHLVTVEGGWPQFGVGAEICARIMEGPAFNFLD 319 Query: 944 APALRVTGADIPMPYAKPLEMEALPRVENVIKSVKKVLHI 1063 APA+RVTGAD+PMPYAK LE ++P+V+++I ++KK L+I Sbjct: 320 APAVRVTGADVPMPYAKILEDNSIPQVKDIIFAIKKTLNI 359
>sp|Q9D051|ODPB_MOUSE Pyruvate dehydrogenase E1 component beta subunit, mitochondrial precursor (PDHE1-B) Length = 359 Score = 469 bits (1208), Expect = e-132 Identities = 225/340 (66%), Positives = 287/340 (84%) Frame = +2 Query: 44 QRHIKLSAAINVKLTVRDALNQALKEEIERDKNVFILGEEVAQYDGAYKVTKGLWKLFGD 223 +R SA V+LTVR+A+NQ + EE+ERD+ VF+LGEEVAQYDGAYKV++GLWK +GD Sbjct: 20 KRRFHRSAPAAVQLTVREAINQGMDEELERDEKVFLLGEEVAQYDGAYKVSRGLWKKYGD 79 Query: 224 ERVVDTPITEMGFAGIAVGAAFHGLRPVCEFMTFNFAMQAVDHIINSAAKGYYMSAGQIK 403 +R++DTPI+EMGFAGIAVGAA GLRP+CEFMTFNF+MQA+D +INSAAK YYMSAG Sbjct: 80 KRIIDTPISEMGFAGIAVGAAMAGLRPICEFMTFNFSMQAIDQVINSAAKTYYMSAGLQP 139 Query: 404 VPIVFRGPNGAAAGVAAQHSQDFSAWFTHCPALKVLSPYSSEDARGLLKSAIRDPDPVIV 583 VPIVFRGPNGA+AGVAAQHSQ F+AW+ HCP LKV+SP++SEDA+GL+KSAIRD +PV++ Sbjct: 140 VPIVFRGPNGASAGVAAQHSQCFAAWYGHCPGLKVVSPWNSEDAKGLIKSAIRDNNPVVM 199 Query: 584 LENEVLYGHPFDTPDEALSPDFLIPIGKAKIEKTGTSVTLVSFSKGVDLCLNAANELAKI 763 LENE++YG F+ P EA S DFLIPIGKAKIE+ GT +T+V+ S+ V CL AA L+K Sbjct: 200 LENELMYGVAFELPAEAQSKDFLIPIGKAKIERQGTHITVVAHSRPVGHCLEAAAVLSKE 259 Query: 764 GVDCEVINLRSLRPLDMETVIKSVVKTHHLVTVEESWPTCGIGSEILARVMETQAFNYLD 943 G++CEVINLR++RP+D+E + SV+KT+HLVTVE WP G+G+EI AR+ME AFN+LD Sbjct: 260 GIECEVINLRTIRPMDIEAIEASVMKTNHLVTVEGGWPQFGVGAEICARIMEGPAFNFLD 319 Query: 944 APALRVTGADIPMPYAKPLEMEALPRVENVIKSVKKVLHI 1063 APA+RVTGAD+PMPYAK LE ++P+V+++I +VKK L+I Sbjct: 320 APAVRVTGADVPMPYAKVLEDNSVPQVKDIIFAVKKTLNI 359
>sp|P49432|ODPB_RAT Pyruvate dehydrogenase E1 component beta subunit, mitochondrial precursor (PDHE1-B) Length = 359 Score = 468 bits (1203), Expect = e-131 Identities = 223/340 (65%), Positives = 286/340 (84%) Frame = +2 Query: 44 QRHIKLSAAINVKLTVRDALNQALKEEIERDKNVFILGEEVAQYDGAYKVTKGLWKLFGD 223 +R SA V+LTVR+A+NQ + EE+ERD+ VF+LGEEVAQYDGAYKV++GLWK +GD Sbjct: 20 KRLFHCSAPAAVQLTVREAINQGMDEELERDEKVFLLGEEVAQYDGAYKVSRGLWKKYGD 79 Query: 224 ERVVDTPITEMGFAGIAVGAAFHGLRPVCEFMTFNFAMQAVDHIINSAAKGYYMSAGQIK 403 +R++DTPI+EMGFAGIAVGAA GLRP+CEFMTFNF+MQA+D +INSAAK YYMSAG Sbjct: 80 KRIIDTPISEMGFAGIAVGAAMAGLRPICEFMTFNFSMQAIDQVINSAAKTYYMSAGLQP 139 Query: 404 VPIVFRGPNGAAAGVAAQHSQDFSAWFTHCPALKVLSPYSSEDARGLLKSAIRDPDPVIV 583 VPIVFRGPNGA+AGVAAQHSQ F+AW+ HCP LKV+SP++SEDA+GL+KSAIRD +PV++ Sbjct: 140 VPIVFRGPNGASAGVAAQHSQCFAAWYGHCPGLKVVSPWNSEDAKGLIKSAIRDDNPVVM 199 Query: 584 LENEVLYGHPFDTPDEALSPDFLIPIGKAKIEKTGTSVTLVSFSKGVDLCLNAANELAKI 763 LENE++YG F+ P EA S DFLIPIGKAKIE+ GT + +V +S+ V CL AA L+K Sbjct: 200 LENELMYGVAFELPTEAQSKDFLIPIGKAKIERQGTHINVVCYSRPVGHCLEAAAVLSKG 259 Query: 764 GVDCEVINLRSLRPLDMETVIKSVVKTHHLVTVEESWPTCGIGSEILARVMETQAFNYLD 943 G++CEVINLR++RP+D+E + SV+KT+HLVTVE WP G+G+EI AR+ME AFN+LD Sbjct: 260 GIECEVINLRTIRPMDIEAIEASVMKTNHLVTVEGGWPQFGVGAEICARIMEGPAFNFLD 319 Query: 944 APALRVTGADIPMPYAKPLEMEALPRVENVIKSVKKVLHI 1063 APA+RVTGAD+PMPYAK LE ++P+V+++I ++KK L+I Sbjct: 320 APAVRVTGADVPMPYAKILEDNSIPQVKDIIFAIKKTLNI 359
>sp|P26269|ODPB_ASCSU Pyruvate dehydrogenase E1 component beta subunit, mitochondrial precursor (PDHE1-B) Length = 361 Score = 464 bits (1194), Expect = e-130 Identities = 220/341 (64%), Positives = 274/341 (80%) Frame = +2 Query: 44 QRHIKLSAAINVKLTVRDALNQALKEEIERDKNVFILGEEVAQYDGAYKVTKGLWKLFGD 223 ++ ++ A+ + +TVRDALN AL EEI+RD VF++GEEVAQYDGAYK++KGLWK +GD Sbjct: 21 EQSVRRLASGTLNVTVRDALNAALDEEIKRDDRVFLIGEEVAQYDGAYKISKGLWKKYGD 80 Query: 224 ERVVDTPITEMGFAGIAVGAAFHGLRPVCEFMTFNFAMQAVDHIINSAAKGYYMSAGQIK 403 R+ DTPITEM AG++VGAA +GLRP+CEFM+ NF+MQ +DHIINSAAK +YMSAG+ Sbjct: 81 GRIWDTPITEMAIAGLSVGAAMNGLRPICEFMSMNFSMQGIDHIINSAAKAHYMSAGRFH 140 Query: 404 VPIVFRGPNGAAAGVAAQHSQDFSAWFTHCPALKVLSPYSSEDARGLLKSAIRDPDPVIV 583 VPIVFRG NGAA GVA QHSQDF+AWF HCP +KV+ PY EDARGLLK+A+RD +PVI Sbjct: 141 VPIVFRGANGAAVGVAQQHSQDFTAWFMHCPGVKVVVPYDCEDARGLLKAAVRDDNPVIC 200 Query: 584 LENEVLYGHPFDTPDEALSPDFLIPIGKAKIEKTGTSVTLVSFSKGVDLCLNAANELAKI 763 LENE+LYG F EA SPDF++P G+AKI++ G +T+VS S GVD+ L+AA+ELAK Sbjct: 201 LENEILYGMKFPVSPEAQSPDFVLPFGQAKIQRPGKDITIVSLSIGVDVSLHAADELAKS 260 Query: 764 GVDCEVINLRSLRPLDMETVIKSVVKTHHLVTVEESWPTCGIGSEILARVMETQAFNYLD 943 G+DCEVINLR +RPLD +TV SV+KT HLVTVE WP CG+G+EI ARV E+ AF YLD Sbjct: 261 GIDCEVINLRCVRPLDFQTVKDSVIKTKHLVTVESGWPNCGVGAEISARVTESDAFGYLD 320 Query: 944 APALRVTGADIPMPYAKPLEMEALPRVENVIKSVKKVLHIQ 1066 P LRVTG D+PMPYA+PLE ALP+ +V+K VKK L++Q Sbjct: 321 GPILRVTGVDVPMPYAQPLETAALPQPADVVKMVKKCLNVQ 361
>sp|P32473|ODPB_YEAST Pyruvate dehydrogenase E1 component beta subunit, mitochondrial precursor (PDHE1-B) Length = 366 Score = 417 bits (1073), Expect = e-116 Identities = 204/329 (62%), Positives = 264/329 (80%), Gaps = 1/329 (0%) Frame = +2 Query: 83 LTVRDALNQALKEEIERDKNVFILGEEVAQYDGAYKVTKGLWKLFGDERVVDTPITEMGF 262 +TVR+ALN A+ EE++RD +VF++GEEVAQY+GAYKV+KGL FG+ RVVDTPITE GF Sbjct: 39 MTVREALNSAMAEELDRDDDVFLIGEEVAQYNGAYKVSKGLLDRFGERRVVDTPITEYGF 98 Query: 263 AGIAVGAAFHGLRPVCEFMTFNFAMQAVDHIINSAAKGYYMSAGQIKVPIVFRGPNGAAA 442 G+AVGAA GL+P+ EFM+FNF+MQA+DH++NSAAK +YMS G K +VFRGPNGAA Sbjct: 99 TGLAVGAALKGLKPIVEFMSFNFSMQAIDHVVNSAAKTHYMSGGTQKCQMVFRGPNGAAV 158 Query: 443 GVAAQHSQDFSAWFTHCPALKVLSPYSSEDARGLLKSAIRDPDPVIVLENEVLYGHPFDT 622 GV AQHSQDFS W+ P LKVL PYS+EDARGLLK+AIRDP+PV+ LENE+LYG F+ Sbjct: 159 GVGAQHSQDFSPWYGSIPGLKVLVPYSAEDARGLLKAAIRDPNPVVFLENELLYGESFEI 218 Query: 623 PDEALSPDFLIPIGKAKIEKTGTSVTLVSFSKGVDLCLNAANEL-AKIGVDCEVINLRSL 799 +EALSP+F +P KAKIE+ GT +++V++++ V L AA L K GV EVINLRS+ Sbjct: 219 SEEALSPEFTLPY-KAKIEREGTDISIVTYTRNVQFSLEAAEILQKKYGVSAEVINLRSI 277 Query: 800 RPLDMETVIKSVVKTHHLVTVEESWPTCGIGSEILARVMETQAFNYLDAPALRVTGADIP 979 RPLD E +IK+V KT+HL+TVE ++P+ G+G+EI+A+VME++AF+YLDAP RVTGAD+P Sbjct: 278 RPLDTEAIIKTVKKTNHLITVESTFPSFGVGAEIVAQVMESEAFDYLDAPIQRVTGADVP 337 Query: 980 MPYAKPLEMEALPRVENVIKSVKKVLHIQ 1066 PYAK LE A P ++K+VK+VL I+ Sbjct: 338 TPYAKELEDFAFPDTPTIVKAVKEVLSIE 366
>sp|Q38799|ODPB_ARATH Pyruvate dehydrogenase E1 component beta subunit, mitochondrial precursor (PDHE1-B) Length = 363 Score = 414 bits (1064), Expect = e-115 Identities = 206/327 (62%), Positives = 255/327 (77%) Frame = +2 Query: 80 KLTVRDALNQALKEEIERDKNVFILGEEVAQYDGAYKVTKGLWKLFGDERVVDTPITEMG 259 ++TVRDALN A+ EE+ D VF++GEEV QY GAYK+TKGL + +G ERV DTPITE G Sbjct: 35 EMTVRDALNSAIDEEMSADPKVFVMGEEVGQYQGAYKITKGLLEKYGPERVYDTPITEAG 94 Query: 260 FAGIAVGAAFHGLRPVCEFMTFNFAMQAVDHIINSAAKGYYMSAGQIKVPIVFRGPNGAA 439 F GI VGAA+ GL+PV EFMTFNF+MQA+DHIINSAAK YMSAGQI VPIVFRGPNGAA Sbjct: 95 FTGIGVGAAYAGLKPVVEFMTFNFSMQAIDHIINSAAKSNYMSAGQINVPIVFRGPNGAA 154 Query: 440 AGVAAQHSQDFSAWFTHCPALKVLSPYSSEDARGLLKSAIRDPDPVIVLENEVLYGHPFD 619 AGV AQHSQ ++AW+ P LKVL+PYS+EDARGLLK+AIRDPDPV+ LENE+LYG F Sbjct: 155 AGVGAQHSQCYAAWYASVPGLKVLAPYSAEDARGLLKAAIRDPDPVVFLENELLYGESFP 214 Query: 620 TPDEALSPDFLIPIGKAKIEKTGTSVTLVSFSKGVDLCLNAANELAKIGVDCEVINLRSL 799 +EAL F +PIGKAKIE+ G VT+V+FSK V L AA +LA+ G+ EVINLRS+ Sbjct: 215 ISEEALDSSFCLPIGKAKIEREGKDVTIVTFSKMVGFALKAAEKLAEEGISAEVINLRSI 274 Query: 800 RPLDMETVIKSVVKTHHLVTVEESWPTCGIGSEILARVMETQAFNYLDAPALRVTGADIP 979 RPLD T+ SV KT LVTVEE +P G+ +EI A V+E ++F+YLDAP R+ GAD+P Sbjct: 275 RPLDRATINASVRKTSRLVTVEEGFPQHGVCAEICASVVE-ESFSYLDAPVERIAGADVP 333 Query: 980 MPYAKPLEMEALPRVENVIKSVKKVLH 1060 MPYA LE ALP++E+++++ K+ + Sbjct: 334 MPYAANLERLALPQIEDIVRASKRACY 360
>sp|Q09171|ODPB_SCHPO Pyruvate dehydrogenase E1 component beta subunit, mitochondrial precursor (PDHE1-B) Length = 366 Score = 405 bits (1041), Expect = e-112 Identities = 198/330 (60%), Positives = 259/330 (78%), Gaps = 1/330 (0%) Frame = +2 Query: 80 KLTVRDALNQALKEEIERDKNVFILGEEVAQYDGAYKVTKGLWKLFGDERVVDTPITEMG 259 ++TVRDALN A++EE++RD VF++GEEVAQY+GAYK+++GL FG +RV+DTPITEMG Sbjct: 37 EMTVRDALNSAMEEEMKRDDRVFLIGEEVAQYNGAYKISRGLLDKFGPKRVIDTPITEMG 96 Query: 260 FAGIAVGAAFHGLRPVCEFMTFNFAMQAVDHIINSAAKGYYMSAGQIKVPIVFRGPNGAA 439 F G+A GAAF GLRP+CEFMTFNF+MQA+DHI+NSAA+ YMS G PIVFRGPNG A Sbjct: 97 FTGLATGAAFAGLRPICEFMTFNFSMQAIDHIVNSAARTLYMSGGIQACPIVFRGPNGPA 156 Query: 440 AGVAAQHSQDFSAWFTHCPALKVLSPYSSEDARGLLKSAIRDPDPVIVLENEVLYGHPFD 619 A VAAQHSQ F+ W+ P LKV+SPYS+EDARGLLK+AIRDP+PV+VLENE+LYG F Sbjct: 157 AAVAAQHSQHFAPWYGSIPGLKVVSPYSAEDARGLLKAAIRDPNPVVVLENEILYGKTFP 216 Query: 620 TPDEALSPDFLIPIGKAKIEKTGTSVTLVSFSKGVDLCLNAANEL-AKIGVDCEVINLRS 796 EALS DF++P G AK+E+ G +T+V S V L AA++L A GV+ EVINLRS Sbjct: 217 ISKEALSEDFVLPFGLAKVERPGKDITIVGESISVVTALEAADKLKADYGVEAEVINLRS 276 Query: 797 LRPLDMETVIKSVVKTHHLVTVEESWPTCGIGSEILARVMETQAFNYLDAPALRVTGADI 976 +RPLD+ T+ SV KT+ +VTV++++ GIGSEI A++ME+ AF+YLDAP RV+ AD+ Sbjct: 277 IRPLDINTIAASVKKTNRIVTVDQAYSQHGIGSEIAAQIMESDAFDYLDAPVERVSMADV 336 Query: 977 PMPYAKPLEMEALPRVENVIKSVKKVLHIQ 1066 PMPY+ P+E ++P + V+ + KK L+I+ Sbjct: 337 PMPYSHPVEAASVPNADVVVAAAKKCLYIK 366
>sp|P52904|ODPB_PEA Pyruvate dehydrogenase E1 component beta subunit, mitochondrial precursor (PDHE1-B) Length = 359 Score = 396 bits (1018), Expect = e-110 Identities = 199/327 (60%), Positives = 245/327 (74%) Frame = +2 Query: 80 KLTVRDALNQALKEEIERDKNVFILGEEVAQYDGAYKVTKGLWKLFGDERVVDTPITEMG 259 ++TVRDALN AL E+ D VF++GEEV +Y GAYKVTKGL + +G ERV+DTPITE G Sbjct: 25 QMTVRDALNSALDVEMSADSKVFLMGEEVGEYQGAYKVTKGLLEKYGPERVLDTPITEAG 84 Query: 260 FAGIAVGAAFHGLRPVCEFMTFNFAMQAVDHIINSAAKGYYMSAGQIKVPIVFRGPNGAA 439 F GI VGAA++GL+PV EFMTFNF+MQA+DHIINSAAK YMSAGQI VPIVFRG NG A Sbjct: 85 FTGIGVGAAYYGLKPVVEFMTFNFSMQAIDHIINSAAKSNYMSAGQISVPIVFRGLNGDA 144 Query: 440 AGVAAQHSQDFSAWFTHCPALKVLSPYSSEDARGLLKSAIRDPDPVIVLENEVLYGHPFD 619 AGV AQHS +++W+ CP LKVL P+S+EDARGLLK+AIRDPDPV+ LENE+LYG F Sbjct: 145 AGVGAQHSHCYASWYGSCPGLKVLVPHSAEDARGLLKAAIRDPDPVVFLENELLYGESFP 204 Query: 620 TPDEALSPDFLIPIGKAKIEKTGTSVTLVSFSKGVDLCLNAANELAKIGVDCEVINLRSL 799 E L F +PIGKAKIE+ G VT+ +FSK V L AA L K G+ EVINLRS+ Sbjct: 205 VSAEVLDSSFWLPIGKAKIEREGKDVTITAFSKMVGFALKAAEILEKEGISAEVINLRSI 264 Query: 800 RPLDMETVIKSVVKTHHLVTVEESWPTCGIGSEILARVMETQAFNYLDAPALRVTGADIP 979 RPLD T+ SV KT+ LVTVEE +P G+G+EI V+E ++F YLDA R+ GAD+P Sbjct: 265 RPLDRPTINASVRKTNRLVTVEEGFPQHGVGAEICTSVIE-ESFGYLDATVERIGGADVP 323 Query: 980 MPYAKPLEMEALPRVENVIKSVKKVLH 1060 MPYA LE +P VE+++++ K+ H Sbjct: 324 MPYAGNLERLVVPHVEDIVRAAKRACH 350
>sp|Q9R9N4|ODPB_RHIME Pyruvate dehydrogenase E1 component, beta subunit Length = 460 Score = 390 bits (1002), Expect = e-108 Identities = 196/326 (60%), Positives = 249/326 (76%) Frame = +2 Query: 83 LTVRDALNQALKEEIERDKNVFILGEEVAQYDGAYKVTKGLWKLFGDERVVDTPITEMGF 262 +TVR+AL A+ EE+ +++VF++GEEVA+Y GAYKVT+GL + FG RVVDTPITE GF Sbjct: 138 MTVREALRDAMAEEMRANEDVFVMGEEVAEYQGAYKVTQGLLQEFGARRVVDTPITEHGF 197 Query: 263 AGIAVGAAFHGLRPVCEFMTFNFAMQAVDHIINSAAKGYYMSAGQIKVPIVFRGPNGAAA 442 AG+ VGAA GLRP+ EFMTFNFAMQA+D IINSAAK YMS GQ+ PIVFRGP+GAAA Sbjct: 198 AGVGVGAAMTGLRPIVEFMTFNFAMQAIDQIINSAAKTLYMSGGQMGAPIVFRGPSGAAA 257 Query: 443 GVAAQHSQDFSAWFTHCPALKVLSPYSSEDARGLLKSAIRDPDPVIVLENEVLYGHPFDT 622 VAAQHSQ ++AW++H P LKV+ PY++ DA+GLLK+AIRDP+PVI LENE+LYG F+ Sbjct: 258 RVAAQHSQCYAAWYSHIPGLKVVMPYTAADAKGLLKAAIRDPNPVIFLENEILYGQSFEV 317 Query: 623 PDEALSPDFLIPIGKAKIEKTGTSVTLVSFSKGVDLCLNAANELAKIGVDCEVINLRSLR 802 P DF++PIGKA+I +TG TLVSF G+ + AA EL G+D E+I+LR++R Sbjct: 318 PK---LDDFVLPIGKARIHRTGKDATLVSFGIGMTYAIKAAAELEAQGIDVEIIDLRTIR 374 Query: 803 PLDMETVIKSVVKTHHLVTVEESWPTCGIGSEILARVMETQAFNYLDAPALRVTGADIPM 982 P+D+ TVI+SV KT LVTVEE +P +G+EI RVM+ QAF+YLDAP L + G D+PM Sbjct: 375 PMDLPTVIESVKKTGRLVTVEEGYPQSSVGTEIATRVMQ-QAFDYLDAPILTIAGKDVPM 433 Query: 983 PYAKPLEMEALPRVENVIKSVKKVLH 1060 PYA LE ALP V V+ +VK V + Sbjct: 434 PYAANLEKLALPNVAEVVDAVKAVCY 459
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 130,672,138
Number of Sequences: 369166
Number of extensions: 2693741
Number of successful extensions: 6693
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 6381
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6619
length of database: 68,354,980
effective HSP length: 113
effective length of database: 47,479,925
effective search space used: 13864138100
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail