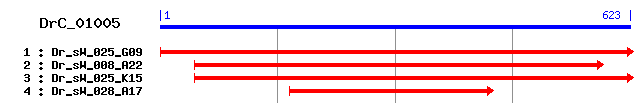
DrC_01005
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_01005 (622 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q04832|HEXP_LEIMA DNA-binding protein HEXBP (Hexamer-bin... 141 1e-33 sp|P53849|GIS2_YEAST Zinc-finger protein GIS2 120 2e-27 sp|P36627|BYR3_SCHPO Cellular nucleic acid binding protein ... 113 3e-25 sp|O42395|CNBP_CHICK Cellular nucleic acid binding protein ... 113 4e-25 sp|P62634|CNBP_RAT Cellular nucleic acid binding protein (C... 112 6e-25 sp|P53996|CNBP_MOUSE Cellular nucleic acid binding protein ... 112 8e-25 sp|Q8WW36|ZCH13_HUMAN Zinc finger CCHC domain containing pr... 100 2e-21 sp|Q966L9|GLH2_CAEEL ATP-dependent RNA helicase glh-2 (Germ... 85 1e-16 sp|O76743|GLH4_CAEEL ATP-dependent RNA helicase glh-4 (Germ... 78 2e-14 sp|P34689|GLH1_CAEEL ATP-dependent RNA helicase glh-1 (Germ... 71 3e-12
>sp|Q04832|HEXP_LEIMA DNA-binding protein HEXBP (Hexamer-binding protein) Length = 271 Score = 141 bits (356), Expect = 1e-33 Identities = 69/183 (37%), Positives = 92/183 (50%), Gaps = 10/183 (5%) Frame = +3 Query: 45 LECYNCGQSGHFSRDCTESKRYGSGARSGGCYNCGENGHISRDCPQNXXXXXXXXXXXXX 224 + C+ CG++GH SRDC S + G+ A+ CY CG+ GH+SRDCP + Sbjct: 70 MTCFRCGEAGHMSRDCPNSAKPGA-AKGFECYKCGQEGHLSRDCPSSQGGSRGGYGQKRG 128 Query: 225 XXXXXXXXXXXXXXXXXXXXXXXXXNNCYNCGQSGHISSYCTQGSRN-NGPSGRGCYKCG 401 CY CG +GHIS C G +G R CYKCG Sbjct: 129 RSGAQGGYSGD--------------RTCYKCGDAGHISRDCPNGQGGYSGAGDRTCYKCG 174 Query: 402 ESGHMARDCVDG-----NEKQVKCYNCNQSGHMSRECPN----GPAEKTCYKCHNTGHIA 554 ++GH++RDC +G KCY C +SGHMSRECP+ G ++ CYKC GHI+ Sbjct: 175 DAGHISRDCPNGQGGYSGAGDRKCYKCGESGHMSRECPSAGSTGSGDRACYKCGKPGHIS 234 Query: 555 RDC 563 R+C Sbjct: 235 REC 237
Score = 138 bits (348), Expect = 1e-32
Identities = 77/200 (38%), Positives = 88/200 (44%), Gaps = 28/200 (14%)
Frame = +3
Query: 48 ECYNCGQSGHFSRDCTES----------KRYGSGARSG-----GCYNCGENGHISRDCPQ 182
ECY CGQ GH SRDC S KR SGA+ G CY CG+ GHISRDCP
Sbjct: 98 ECYKCGQEGHLSRDCPSSQGGSRGGYGQKRGRSGAQGGYSGDRTCYKCGDAGHISRDCPN 157
Query: 183 NXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXNNCYNCGQSGHISSYCTQGSR 362
CY CG +GHIS C G
Sbjct: 158 GQGGYSGAGD-----------------------------RTCYKCGDAGHISRDCPNGQG 188
Query: 363 N-NGPSGRGCYKCGESGHMARDCVDGNEK---QVKCYNCNQSGHMSRECP---------N 503
+G R CYKCGESGHM+R+C CY C + GH+SRECP
Sbjct: 189 GYSGAGDRKCYKCGESGHMSRECPSAGSTGSGDRACYKCGKPGHISRECPEAGGSYGGSR 248
Query: 504 GPAEKTCYKCHNTGHIARDC 563
G ++TCYKC GHI+RDC
Sbjct: 249 GGGDRTCYKCGEAGHISRDC 268
Score = 127 bits (319), Expect = 2e-29
Identities = 63/197 (31%), Positives = 87/197 (44%), Gaps = 26/197 (13%)
Frame = +3
Query: 51 CYNCGQSGHFSRDCTESKRYGSGARSGGCYNCGENGHISRDCPQNXXXXXXXXXXXXXXX 230
C NCG+ GH++R+C E+ G RS C+ CGE GH+SR+CP
Sbjct: 18 CRNCGKEGHYARECPEADSKGD-ERSTTCFRCGEEGHMSRECPNEARSGAAGAM------ 70
Query: 231 XXXXXXXXXXXXXXXXXXXXXXXNNCYNCGQSGHISSYCTQGSRNNGPSGRGCYKCGESG 410
C+ CG++GH+S C ++ G CYKCG+ G
Sbjct: 71 ------------------------TCFRCGEAGHMSRDCPNSAKPGAAKGFECYKCGQEG 106
Query: 411 HMARDCVD--------------------GNEKQVKCYNCNQSGHMSRECPN------GPA 512
H++RDC G CY C +GH+SR+CPN G
Sbjct: 107 HLSRDCPSSQGGSRGGYGQKRGRSGAQGGYSGDRTCYKCGDAGHISRDCPNGQGGYSGAG 166
Query: 513 EKTCYKCHNTGHIARDC 563
++TCYKC + GHI+RDC
Sbjct: 167 DRTCYKCGDAGHISRDC 183
Score = 103 bits (258), Expect = 3e-22
Identities = 53/182 (29%), Positives = 76/182 (41%), Gaps = 25/182 (13%)
Frame = +3
Query: 93 TESKRYGSGARSGGCYNCGENGHISRDCPQNXXXXXXXXXXXXXXXXXXXXXXXXXXXXX 272
TE + S C NCG+ GH +R+CP+
Sbjct: 4 TEDVKRPRTESSTSCRNCGKEGHYARECPE------------------------------ 33
Query: 273 XXXXXXXXXNNCYNCGQSGHISSYCTQGSRNNGPSGRGCYKCGESGHMARDCVD----GN 440
C+ CG+ GH+S C +R+ C++CGE+GHM+RDC + G
Sbjct: 34 ADSKGDERSTTCFRCGEEGHMSRECPNEARSGAAGAMTCFRCGEAGHMSRDCPNSAKPGA 93
Query: 441 EKQVKCYNCNQSGHMSRECPNG---------------------PAEKTCYKCHNTGHIAR 557
K +CY C Q GH+SR+CP+ ++TCYKC + GHI+R
Sbjct: 94 AKGFECYKCGQEGHLSRDCPSSQGGSRGGYGQKRGRSGAQGGYSGDRTCYKCGDAGHISR 153
Query: 558 DC 563
DC
Sbjct: 154 DC 155
Score = 64.3 bits (155), Expect = 2e-10
Identities = 27/48 (56%), Positives = 32/48 (66%), Gaps = 3/48 (6%)
Frame = +3
Query: 51 CYNCGQSGHFSRDCTESKRYGSGARSGG---CYNCGENGHISRDCPQN 185
CY CG+ GH SR+C E+ G+R GG CY CGE GHISRDCP +
Sbjct: 224 CYKCGKPGHISRECPEAGGSYGGSRGGGDRTCYKCGEAGHISRDCPSS 271
Score = 47.0 bits (110), Expect = 4e-05
Identities = 17/49 (34%), Positives = 28/49 (57%), Gaps = 5/49 (10%)
Frame = +3
Query: 441 EKQVKCYNCNQSGHMSRECPNGPAE-----KTCYKCHNTGHIARDCHDD 572
E C NC + GH +RECP ++ TC++C GH++R+C ++
Sbjct: 13 ESSTSCRNCGKEGHYARECPEADSKGDERSTTCFRCGEEGHMSRECPNE 61
>sp|P53849|GIS2_YEAST Zinc-finger protein GIS2 Length = 153 Score = 120 bits (302), Expect = 2e-27 Identities = 62/181 (34%), Positives = 82/181 (45%), Gaps = 1/181 (0%) Frame = +3 Query: 24 EQMSQRRLECYNCGQSGHFSRDCTESKRYGSGARSGGCYNCGENGHISRDCPQNXXXXXX 203 E RL CYNC + GH DCT + CYNCGE GH+ +C Sbjct: 17 EDCDSERL-CYNCNKPGHVQTDCTMPRT----VEFKQCYNCGETGHVRSECT-------- 63 Query: 204 XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXNNCYNCGQSGHISSYCTQGSRNNGPSGR 383 C+NC Q+GHIS C + + + S Sbjct: 64 -------------------------------VQRCFNCNQTGHISRECPEPKKTSRFSKV 92 Query: 384 GCYKCGESGHMARDCV-DGNEKQVKCYNCNQSGHMSRECPNGPAEKTCYKCHNTGHIARD 560 CYKCG HMA+DC+ + +KCY C Q+GHMSR+C N ++ CY C+ TGHI++D Sbjct: 93 SCYKCGGPNHMAKDCMKEDGISGLKCYTCGQAGHMSRDCQN---DRLCYNCNETGHISKD 149 Query: 561 C 563 C Sbjct: 150 C 150
Score = 100 bits (248), Expect = 4e-21
Identities = 48/151 (31%), Positives = 71/151 (47%)
Frame = +3
Query: 48 ECYNCGQSGHFSRDCTESKRYGSGARSGGCYNCGENGHISRDCPQNXXXXXXXXXXXXXX 227
+CYNCG++GH +CT + C+NC + GHISR+CP+
Sbjct: 48 QCYNCGETGHVRSECTVQR----------CFNCNQTGHISRECPEPKKTSRFSKV----- 92
Query: 228 XXXXXXXXXXXXXXXXXXXXXXXXNNCYNCGQSGHISSYCTQGSRNNGPSGRGCYKCGES 407
+CY CG H++ C + +G SG CY CG++
Sbjct: 93 -------------------------SCYKCGGPNHMAKDCM---KEDGISGLKCYTCGQA 124
Query: 408 GHMARDCVDGNEKQVKCYNCNQSGHMSRECP 500
GHM+RDC + CYNCN++GH+S++CP
Sbjct: 125 GHMSRDC----QNDRLCYNCNETGHISKDCP 151
Score = 98.6 bits (244), Expect = 1e-20
Identities = 55/192 (28%), Positives = 84/192 (43%), Gaps = 6/192 (3%)
Frame = +3
Query: 30 MSQRRLECYNCGQSGHFSRDCTESKRYGSGARSGGCYNCGENGHISRDCPQNXXXXXXXX 209
MSQ+ CY CG+ GH + DC +S+R CYNC + GH+ DC
Sbjct: 1 MSQKA--CYVCGKIGHLAEDC-DSERL--------CYNCNKPGHVQTDCTM--------- 40
Query: 210 XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXNNCYNCGQSGHISSYCTQGSRNNGPSGRGC 389
CYNCG++GH+ S CT + C
Sbjct: 41 ------------------------PRTVEFKQCYNCGETGHVRSECTV---------QRC 67
Query: 390 YKCGESGHMARDCVDGNE----KQVKCYNCNQSGHMSREC--PNGPAEKTCYKCHNTGHI 551
+ C ++GH++R+C + + +V CY C HM+++C +G + CY C GH+
Sbjct: 68 FNCNQTGHISRECPEPKKTSRFSKVSCYKCGGPNHMAKDCMKEDGISGLKCYTCGQAGHM 127
Query: 552 ARDCHDD*F*FN 587
+RDC +D +N
Sbjct: 128 SRDCQNDRLCYN 139
Score = 59.3 bits (142), Expect = 8e-09
Identities = 24/46 (52%), Positives = 29/46 (63%)
Frame = +3
Query: 45 LECYNCGQSGHFSRDCTESKRYGSGARSGGCYNCGENGHISRDCPQ 182
L+CY CGQ+GH SRDC + CYNC E GHIS+DCP+
Sbjct: 116 LKCYTCGQAGHMSRDCQNDRL---------CYNCNETGHISKDCPK 152
Score = 31.6 bits (70), Expect = 1.7
Identities = 9/17 (52%), Positives = 14/17 (82%)
Frame = +3
Query: 51 CYNCGQSGHFSRDCTES 101
CYNC ++GH S+DC ++
Sbjct: 137 CYNCNETGHISKDCPKA 153
>sp|P36627|BYR3_SCHPO Cellular nucleic acid binding protein homolog Length = 179 Score = 113 bits (283), Expect = 3e-25 Identities = 62/177 (35%), Positives = 79/177 (44%), Gaps = 6/177 (3%) Frame = +3 Query: 51 CYNCGQSGHFSRDCTESKRYGSGARSGGCYNCGENGHISRDCPQNXXXXXXXXXXXXXXX 230 CYNC Q+GH + +CTE ++ + CY CG GH+ RDCP + Sbjct: 38 CYNCNQTGHKASECTEPQQEKT------CYACGTAGHLVRDCPSSPNPRQGA-------- 83 Query: 231 XXXXXXXXXXXXXXXXXXXXXXXNNCYNCGQSGHISSYC-TQGSRNNGPSG-----RGCY 392 CY CG+ GHI+ C T G ++ G G CY Sbjct: 84 ------------------------ECYKCGRVGHIARDCRTNGQQSGGRFGGHRSNMNCY 119 Query: 393 KCGESGHMARDCVDGNEKQVKCYNCNQSGHMSRECPNGPAEKTCYKCHNTGHIARDC 563 CG GH ARDC G VKCY+C + GH S EC + CYKC+ GHIA +C Sbjct: 120 ACGSYGHQARDCTMG----VKCYSCGKIGHRSFECQQASDGQLCYKCNQPGHIAVNC 172
Score = 89.7 bits (221), Expect = 5e-18
Identities = 38/89 (42%), Positives = 54/89 (60%), Gaps = 3/89 (3%)
Frame = +3
Query: 306 CYNCGQSGHISSYCTQGSRNNGPSGRGCYKCGESGHMARDCVDGNEKQVKCYNCNQSGHM 485
CYNCG++GH + CT+GS CY C ++GH A +C + +++ CY C +GH+
Sbjct: 19 CYNCGENGHQARECTKGSI--------CYNCNQTGHKASECTEPQQEKT-CYACGTAGHL 69
Query: 486 SRECPNGPAEK---TCYKCHNTGHIARDC 563
R+CP+ P + CYKC GHIARDC
Sbjct: 70 VRDCPSSPNPRQGAECYKCGRVGHIARDC 98
Score = 89.7 bits (221), Expect = 5e-18
Identities = 50/154 (32%), Positives = 63/154 (40%)
Frame = +3
Query: 36 QRRLECYNCGQSGHFSRDCTESKRYGSGARSGGCYNCGENGHISRDCPQNXXXXXXXXXX 215
Q+ CY CG +GH RDC S GA CY CG GHI+RDC N
Sbjct: 55 QQEKTCYACGTAGHLVRDCPSSPNPRQGAE---CYKCGRVGHIARDCRTNGQQSGGRFGG 111
Query: 216 XXXXXXXXXXXXXXXXXXXXXXXXXXXXNNCYNCGQSGHISSYCTQGSRNNGPSGRGCYK 395
NCY CG GH + CT G + CY
Sbjct: 112 HRSNM------------------------NCYACGSYGHQARDCTMGVK--------CYS 139
Query: 396 CGESGHMARDCVDGNEKQVKCYNCNQSGHMSREC 497
CG+ GH + +C ++ Q+ CY CNQ GH++ C
Sbjct: 140 CGKIGHRSFECQQASDGQL-CYKCNQPGHIAVNC 172
Score = 81.3 bits (199), Expect = 2e-15
Identities = 33/62 (53%), Positives = 39/62 (62%)
Frame = +3
Query: 378 GRGCYKCGESGHMARDCVDGNEKQVKCYNCNQSGHMSRECPNGPAEKTCYKCHNTGHIAR 557
G CY CGE+GH AR+C G+ CYNCNQ+GH + EC EKTCY C GH+ R
Sbjct: 16 GPRCYNCGENGHQARECTKGSI----CYNCNQTGHKASECTEPQQEKTCYACGTAGHLVR 71
Query: 558 DC 563
DC
Sbjct: 72 DC 73
>sp|O42395|CNBP_CHICK Cellular nucleic acid binding protein (CNBP) (Zinc finger protein 9) Length = 172 Score = 113 bits (282), Expect = 4e-25 Identities = 58/186 (31%), Positives = 84/186 (45%), Gaps = 14/186 (7%) Frame = +3 Query: 48 ECYNCGQSGHFSRDCTESKRYGSGARSGG--------------CYNCGENGHISRDCPQN 185 EC+ CG++GH++R+C G G RS G CY CGE+GH+++DC Sbjct: 5 ECFKCGRTGHWARECPTGIGRGRGMRSRGRAGFQFMSSSLPDICYRCGESGHLAKDCDLQ 64 Query: 186 XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXNNCYNCGQSGHISSYCTQGSRN 365 CYNCG+ GHI+ C + R Sbjct: 65 EDKA------------------------------------CYNCGRGGHIAKDCKEPKRE 88 Query: 366 NGPSGRGCYKCGESGHMARDCVDGNEKQVKCYNCNQSGHMSRECPNGPAEKTCYKCHNTG 545 CY CG+ GH+ARDC +E+ KCY+C + GH+ ++C + CY+C TG Sbjct: 89 REQC---CYNCGKPGHLARDCDHADEQ--KCYSCGEFGHIQKDC----TKVKCYRCGETG 139 Query: 546 HIARDC 563 H+A +C Sbjct: 140 HVAINC 145
Score = 101 bits (252), Expect = 1e-21
Identities = 52/149 (34%), Positives = 69/149 (46%)
Frame = +3
Query: 51 CYNCGQSGHFSRDCTESKRYGSGARSGGCYNCGENGHISRDCPQNXXXXXXXXXXXXXXX 230
CYNCG+ GH ++DC E KR R CYNCG+ GH++RDC
Sbjct: 69 CYNCGRGGHIAKDCKEPKR----EREQCCYNCGKPGHLARDCDH---------------- 108
Query: 231 XXXXXXXXXXXXXXXXXXXXXXXNNCYNCGQSGHISSYCTQGSRNNGPSGRGCYKCGESG 410
CY+CG+ GHI CT+ CY+CGE+G
Sbjct: 109 --------------------ADEQKCYSCGEFGHIQKDCTKVK---------CYRCGETG 139
Query: 411 HMARDCVDGNEKQVKCYNCNQSGHMSREC 497
H+A +C +E V CY C +SGH++REC
Sbjct: 140 HVAINCSKTSE--VNCYRCGESGHLAREC 166
Score = 89.7 bits (221), Expect = 5e-18
Identities = 44/147 (29%), Positives = 65/147 (44%), Gaps = 1/147 (0%)
Frame = +3
Query: 126 SGGCYNCGENGHISRDCPQNXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXNN 305
S C+ CG GH +R+CP +
Sbjct: 3 SNECFKCGRTGHWARECPTG---------------IGRGRGMRSRGRAGFQFMSSSLPDI 47
Query: 306 CYNCGQSGHISSYCTQGSRNNGPSGRGCYKCGESGHMARDCVD-GNEKQVKCYNCNQSGH 482
CY CG+SGH++ C + + CY CG GH+A+DC + E++ CYNC + GH
Sbjct: 48 CYRCGESGHLAKDC------DLQEDKACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGH 101
Query: 483 MSRECPNGPAEKTCYKCHNTGHIARDC 563
++R+C + +K CY C GHI +DC
Sbjct: 102 LARDCDHADEQK-CYSCGEFGHIQKDC 127
Score = 80.1 bits (196), Expect = 4e-15
Identities = 44/138 (31%), Positives = 59/138 (42%)
Frame = +3
Query: 15 DFFEQMSQRRLECYNCGQSGHFSRDCTESKRYGSGARSGGCYNCGENGHISRDCPQNXXX 194
D E +R CYNCG+ GH +RDC A CY+CGE GHI +DC +
Sbjct: 81 DCKEPKREREQCCYNCGKPGHLARDCDH-------ADEQKCYSCGEFGHIQKDCTK---- 129
Query: 195 XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXNNCYNCGQSGHISSYCTQGSRNNGP 374
CY CG++GH++ C++ S N
Sbjct: 130 -----------------------------------VKCYRCGETGHVAINCSKTSEVN-- 152
Query: 375 SGRGCYKCGESGHMARDC 428
CY+CGESGH+AR+C
Sbjct: 153 ----CYRCGESGHLAREC 166
Score = 60.5 bits (145), Expect = 3e-09
Identities = 28/84 (33%), Positives = 41/84 (48%), Gaps = 19/84 (22%)
Frame = +3
Query: 375 SGRGCYKCGESGHMARDCVDG-------------------NEKQVKCYNCNQSGHMSREC 497
S C+KCG +GH AR+C G + CY C +SGH++++C
Sbjct: 2 SSNECFKCGRTGHWARECPTGIGRGRGMRSRGRAGFQFMSSSLPDICYRCGESGHLAKDC 61
Query: 498 PNGPAEKTCYKCHNTGHIARDCHD 569
+ +K CY C GHIA+DC +
Sbjct: 62 -DLQEDKACYNCGRGGHIAKDCKE 84
Score = 33.1 bits (74), Expect = 0.58
Identities = 10/17 (58%), Positives = 14/17 (82%)
Frame = +3
Query: 45 LECYNCGQSGHFSRDCT 95
+ CY CG+SGH +R+CT
Sbjct: 151 VNCYRCGESGHLARECT 167
>sp|P62634|CNBP_RAT Cellular nucleic acid binding protein (CNBP) (Zinc finger protein 9) sp|P62633|CNBP_HUMAN Cellular nucleic acid binding protein (CNBP) (Zinc finger protein 9) Length = 177 Score = 112 bits (281), Expect = 6e-25 Identities = 59/192 (30%), Positives = 85/192 (44%), Gaps = 20/192 (10%) Frame = +3 Query: 48 ECYNCGQSGHFSRDCTESKRYGSGARSGG--------------------CYNCGENGHIS 167 EC+ CG+SGH++R+C G G RS G CY CGE+GH++ Sbjct: 5 ECFKCGRSGHWARECPTGGGRGRGMRSRGRGGFTSDRGFQFVSSSLPDICYRCGESGHLA 64 Query: 168 RDCPQNXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXNNCYNCGQSGHISSYC 347 +DC + CYNCG+ GHI+ C Sbjct: 65 KDCDLQE-------------------------------------DACYNCGRGGHIAKDC 87 Query: 348 TQGSRNNGPSGRGCYKCGESGHMARDCVDGNEKQVKCYNCNQSGHMSRECPNGPAEKTCY 527 + R CY CG+ GH+ARDC +E+ KCY+C + GH+ ++C + CY Sbjct: 88 KEPKREREQC---CYNCGKPGHLARDCDHADEQ--KCYSCGEFGHIQKDC----TKVKCY 138 Query: 528 KCHNTGHIARDC 563 +C TGH+A +C Sbjct: 139 RCGETGHVAINC 150
Score = 101 bits (252), Expect = 1e-21
Identities = 52/149 (34%), Positives = 69/149 (46%)
Frame = +3
Query: 51 CYNCGQSGHFSRDCTESKRYGSGARSGGCYNCGENGHISRDCPQNXXXXXXXXXXXXXXX 230
CYNCG+ GH ++DC E KR R CYNCG+ GH++RDC
Sbjct: 74 CYNCGRGGHIAKDCKEPKR----EREQCCYNCGKPGHLARDCDH---------------- 113
Query: 231 XXXXXXXXXXXXXXXXXXXXXXXNNCYNCGQSGHISSYCTQGSRNNGPSGRGCYKCGESG 410
CY+CG+ GHI CT+ CY+CGE+G
Sbjct: 114 --------------------ADEQKCYSCGEFGHIQKDCTKVK---------CYRCGETG 144
Query: 411 HMARDCVDGNEKQVKCYNCNQSGHMSREC 497
H+A +C +E V CY C +SGH++REC
Sbjct: 145 HVAINCSKTSE--VNCYRCGESGHLAREC 171
Score = 89.7 bits (221), Expect = 5e-18
Identities = 43/111 (38%), Positives = 59/111 (53%), Gaps = 23/111 (20%)
Frame = +3
Query: 300 NNCYNCGQSGHISSYC-TQGSRNNGPSGRG--------------------CYKCGESGHM 416
N C+ CG+SGH + C T G R G RG CY+CGESGH+
Sbjct: 4 NECFKCGRSGHWARECPTGGGRGRGMRSRGRGGFTSDRGFQFVSSSLPDICYRCGESGHL 63
Query: 417 ARDCVDGNEKQVKCYNCNQSGHMSREC--PNGPAEKTCYKCHNTGHIARDC 563
A+DC + ++ CYNC + GH++++C P E+ CY C GH+ARDC
Sbjct: 64 AKDC---DLQEDACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGHLARDC 111
Score = 88.2 bits (217), Expect = 2e-17
Identities = 44/147 (29%), Positives = 64/147 (43%), Gaps = 1/147 (0%)
Frame = +3
Query: 126 SGGCYNCGENGHISRDCPQNXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXNN 305
S C+ CG +GH +R+CP +
Sbjct: 3 SNECFKCGRSGHWARECPTG---------GGRGRGMRSRGRGGFTSDRGFQFVSSSLPDI 53
Query: 306 CYNCGQSGHISSYCTQGSRNNGPSGRGCYKCGESGHMARDCVD-GNEKQVKCYNCNQSGH 482
CY CG+SGH++ C CY CG GH+A+DC + E++ CYNC + GH
Sbjct: 54 CYRCGESGHLAKDCDL-------QEDACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGH 106
Query: 483 MSRECPNGPAEKTCYKCHNTGHIARDC 563
++R+C + +K CY C GHI +DC
Sbjct: 107 LARDCDHADEQK-CYSCGEFGHIQKDC 132
Score = 80.1 bits (196), Expect = 4e-15
Identities = 44/138 (31%), Positives = 59/138 (42%)
Frame = +3
Query: 15 DFFEQMSQRRLECYNCGQSGHFSRDCTESKRYGSGARSGGCYNCGENGHISRDCPQNXXX 194
D E +R CYNCG+ GH +RDC A CY+CGE GHI +DC +
Sbjct: 86 DCKEPKREREQCCYNCGKPGHLARDCDH-------ADEQKCYSCGEFGHIQKDCTK---- 134
Query: 195 XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXNNCYNCGQSGHISSYCTQGSRNNGP 374
CY CG++GH++ C++ S N
Sbjct: 135 -----------------------------------VKCYRCGETGHVAINCSKTSEVN-- 157
Query: 375 SGRGCYKCGESGHMARDC 428
CY+CGESGH+AR+C
Sbjct: 158 ----CYRCGESGHLAREC 171
Score = 58.2 bits (139), Expect = 2e-08
Identities = 29/90 (32%), Positives = 39/90 (43%), Gaps = 25/90 (27%)
Frame = +3
Query: 375 SGRGCYKCGESGHMARDCVDGNEK-------------------------QVKCYNCNQSG 479
S C+KCG SGH AR+C G + CY C +SG
Sbjct: 2 SSNECFKCGRSGHWARECPTGGGRGRGMRSRGRGGFTSDRGFQFVSSSLPDICYRCGESG 61
Query: 480 HMSRECPNGPAEKTCYKCHNTGHIARDCHD 569
H++++C E CY C GHIA+DC +
Sbjct: 62 HLAKDC--DLQEDACYNCGRGGHIAKDCKE 89
Score = 33.1 bits (74), Expect = 0.58
Identities = 10/17 (58%), Positives = 14/17 (82%)
Frame = +3
Query: 45 LECYNCGQSGHFSRDCT 95
+ CY CG+SGH +R+CT
Sbjct: 156 VNCYRCGESGHLARECT 172
>sp|P53996|CNBP_MOUSE Cellular nucleic acid binding protein (CNBP) (Zinc finger protein 9) Length = 178 Score = 112 bits (280), Expect = 8e-25 Identities = 59/192 (30%), Positives = 84/192 (43%), Gaps = 20/192 (10%) Frame = +3 Query: 48 ECYNCGQSGHFSRDCTESKRYGSGARSGG--------------------CYNCGENGHIS 167 EC+ CG+SGH++R+C G G RS G CY CGE+GH++ Sbjct: 5 ECFKCGRSGHWARECPTGGGRGRGMRSRGRGGFTSDRGFQFVSSSLPDICYRCGESGHLA 64 Query: 168 RDCPQNXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXNNCYNCGQSGHISSYC 347 +DC CYNCG+ GHI+ C Sbjct: 65 KDCDLQEDEA------------------------------------CYNCGRGGHIAKDC 88 Query: 348 TQGSRNNGPSGRGCYKCGESGHMARDCVDGNEKQVKCYNCNQSGHMSRECPNGPAEKTCY 527 + R CY CG+ GH+ARDC +E+ KCY+C + GH+ ++C + CY Sbjct: 89 KEPKREREQC---CYNCGKPGHLARDCDHADEQ--KCYSCGEFGHIQKDC----TKVKCY 139 Query: 528 KCHNTGHIARDC 563 +C TGH+A +C Sbjct: 140 RCGETGHVAINC 151
Score = 101 bits (252), Expect = 1e-21
Identities = 52/149 (34%), Positives = 69/149 (46%)
Frame = +3
Query: 51 CYNCGQSGHFSRDCTESKRYGSGARSGGCYNCGENGHISRDCPQNXXXXXXXXXXXXXXX 230
CYNCG+ GH ++DC E KR R CYNCG+ GH++RDC
Sbjct: 75 CYNCGRGGHIAKDCKEPKR----EREQCCYNCGKPGHLARDCDH---------------- 114
Query: 231 XXXXXXXXXXXXXXXXXXXXXXXNNCYNCGQSGHISSYCTQGSRNNGPSGRGCYKCGESG 410
CY+CG+ GHI CT+ CY+CGE+G
Sbjct: 115 --------------------ADEQKCYSCGEFGHIQKDCTKVK---------CYRCGETG 145
Query: 411 HMARDCVDGNEKQVKCYNCNQSGHMSREC 497
H+A +C +E V CY C +SGH++REC
Sbjct: 146 HVAINCSKTSE--VNCYRCGESGHLAREC 172
Score = 90.1 bits (222), Expect = 4e-18
Identities = 45/111 (40%), Positives = 59/111 (53%), Gaps = 23/111 (20%)
Frame = +3
Query: 300 NNCYNCGQSGHISSYC-TQGSRNNGPSGRG--------------------CYKCGESGHM 416
N C+ CG+SGH + C T G R G RG CY+CGESGH+
Sbjct: 4 NECFKCGRSGHWARECPTGGGRGRGMRSRGRGGFTSDRGFQFVSSSLPDICYRCGESGHL 63
Query: 417 ARDCVDGNEKQVKCYNCNQSGHMSREC--PNGPAEKTCYKCHNTGHIARDC 563
A+DC D E + CYNC + GH++++C P E+ CY C GH+ARDC
Sbjct: 64 AKDC-DLQEDEA-CYNCGRGGHIAKDCKEPKREREQCCYNCGKPGHLARDC 112
Score = 89.4 bits (220), Expect = 7e-18
Identities = 44/147 (29%), Positives = 65/147 (44%), Gaps = 1/147 (0%)
Frame = +3
Query: 126 SGGCYNCGENGHISRDCPQNXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXNN 305
S C+ CG +GH +R+CP +
Sbjct: 3 SNECFKCGRSGHWARECPTG---------GGRGRGMRSRGRGGFTSDRGFQFVSSSLPDI 53
Query: 306 CYNCGQSGHISSYCTQGSRNNGPSGRGCYKCGESGHMARDCVD-GNEKQVKCYNCNQSGH 482
CY CG+SGH++ C + CY CG GH+A+DC + E++ CYNC + GH
Sbjct: 54 CYRCGESGHLAKDC------DLQEDEACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGH 107
Query: 483 MSRECPNGPAEKTCYKCHNTGHIARDC 563
++R+C + +K CY C GHI +DC
Sbjct: 108 LARDCDHADEQK-CYSCGEFGHIQKDC 133
Score = 80.1 bits (196), Expect = 4e-15
Identities = 44/138 (31%), Positives = 59/138 (42%)
Frame = +3
Query: 15 DFFEQMSQRRLECYNCGQSGHFSRDCTESKRYGSGARSGGCYNCGENGHISRDCPQNXXX 194
D E +R CYNCG+ GH +RDC A CY+CGE GHI +DC +
Sbjct: 87 DCKEPKREREQCCYNCGKPGHLARDCDH-------ADEQKCYSCGEFGHIQKDCTK---- 135
Query: 195 XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXNNCYNCGQSGHISSYCTQGSRNNGP 374
CY CG++GH++ C++ S N
Sbjct: 136 -----------------------------------VKCYRCGETGHVAINCSKTSEVN-- 158
Query: 375 SGRGCYKCGESGHMARDC 428
CY+CGESGH+AR+C
Sbjct: 159 ----CYRCGESGHLAREC 172
Score = 58.2 bits (139), Expect = 2e-08
Identities = 28/90 (31%), Positives = 41/90 (45%), Gaps = 25/90 (27%)
Frame = +3
Query: 375 SGRGCYKCGESGHMARDCVDGNEK-------------------------QVKCYNCNQSG 479
S C+KCG SGH AR+C G + CY C +SG
Sbjct: 2 SSNECFKCGRSGHWARECPTGGGRGRGMRSRGRGGFTSDRGFQFVSSSLPDICYRCGESG 61
Query: 480 HMSRECPNGPAEKTCYKCHNTGHIARDCHD 569
H++++C + ++ CY C GHIA+DC +
Sbjct: 62 HLAKDC-DLQEDEACYNCGRGGHIAKDCKE 90
Score = 33.1 bits (74), Expect = 0.58
Identities = 10/17 (58%), Positives = 14/17 (82%)
Frame = +3
Query: 45 LECYNCGQSGHFSRDCT 95
+ CY CG+SGH +R+CT
Sbjct: 157 VNCYRCGESGHLARECT 173
>sp|Q8WW36|ZCH13_HUMAN Zinc finger CCHC domain containing protein 13 Length = 166 Score = 100 bits (250), Expect = 2e-21 Identities = 58/186 (31%), Positives = 81/186 (43%), Gaps = 16/186 (8%) Frame = +3 Query: 54 YNCGQSGHFSRDCTESKRYGSGARSGG----------------CYNCGENGHISRDCPQN 185 + CG SGH++R C R G+G R GG CY CGE+G +++C Sbjct: 7 FACGHSGHWARGCP---RGGAGGRRGGGHGRGSQCGSTTLSYTCYCCGESGRNAKNC--- 60 Query: 186 XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXNNCYNCGQSGHISSYCTQGSRN 365 N CYNCG+SGHI+ C R Sbjct: 61 ----------------------------------VLLGNICYNCGRSGHIAKDCKDPKRE 86 Query: 366 NGPSGRGCYKCGESGHMARDCVDGNEKQVKCYNCNQSGHMSRECPNGPAEKTCYKCHNTG 545 + CY CG GH+ARDC +K+ KCY+C + GH+ ++C A+ CY+C G Sbjct: 87 R---RQHCYTCGRLGHLARDC--DRQKEQKCYSCGKLGHIQKDC----AQVKCYRCGEIG 137 Query: 546 HIARDC 563 H+A +C Sbjct: 138 HVAINC 143
Score = 76.6 bits (187), Expect = 5e-14
Identities = 41/149 (27%), Positives = 59/149 (39%)
Frame = +3
Query: 51 CYNCGQSGHFSRDCTESKRYGSGARSGGCYNCGENGHISRDCPQNXXXXXXXXXXXXXXX 230
CY CG+SG +++C CYNCG +GHI++DC
Sbjct: 47 CYCCGESGRNAKNCV--------LLGNICYNCGRSGHIAKDCKD---------------- 82
Query: 231 XXXXXXXXXXXXXXXXXXXXXXXNNCYNCGQSGHISSYCTQGSRNNGPSGRGCYKCGESG 410
+CY CG+ GH++ C + CY CG+ G
Sbjct: 83 -----------------PKRERRQHCYTCGRLGHLARDCDRQKEQK------CYSCGKLG 119
Query: 411 HMARDCVDGNEKQVKCYNCNQSGHMSREC 497
H+ +DC QVKCY C + GH++ C
Sbjct: 120 HIQKDCA-----QVKCYRCGEIGHVAINC 143
Score = 54.7 bits (130), Expect = 2e-07
Identities = 29/106 (27%), Positives = 40/106 (37%)
Frame = +3
Query: 36 QRRLECYNCGQSGHFSRDCTESKRYGSGARSGGCYNCGENGHISRDCPQNXXXXXXXXXX 215
+RR CY CG+ GH +RDC K CY+CG+ GHI +DC Q
Sbjct: 86 ERRQHCYTCGRLGHLARDCDRQKEQ-------KCYSCGKLGHIQKDCAQ----------- 127
Query: 216 XXXXXXXXXXXXXXXXXXXXXXXXXXXXNNCYNCGQSGHISSYCTQ 353
CY CG+ GH++ C++
Sbjct: 128 ----------------------------VKCYRCGEIGHVAINCSK 145
Score = 51.2 bits (121), Expect = 2e-06
Identities = 30/90 (33%), Positives = 40/90 (44%), Gaps = 3/90 (3%)
Frame = +3
Query: 309 YNCGQSGHISSYCTQGS---RNNGPSGRGCYKCGESGHMARDCVDGNEKQVKCYNCNQSG 479
+ CG SGH + C +G R G GRG +CG + CY C +SG
Sbjct: 7 FACGHSGHWARGCPRGGAGGRRGGGHGRGS-QCGST-----------TLSYTCYCCGESG 54
Query: 480 HMSRECPNGPAEKTCYKCHNTGHIARDCHD 569
++ C CY C +GHIA+DC D
Sbjct: 55 RNAKNCVL--LGNICYNCGRSGHIAKDCKD 82
>sp|Q966L9|GLH2_CAEEL ATP-dependent RNA helicase glh-2 (Germline helicase 2) Length = 974 Score = 85.1 bits (209), Expect = 1e-16 Identities = 57/209 (27%), Positives = 75/209 (35%), Gaps = 38/209 (18%) Frame = +3 Query: 51 CYNCGQSGHFSRDCTESK--RYGSGARSGGC--YNCGENGHISRDCPQNXXXXXXXXXXX 218 CYNC Q GH SRDC E + R G +GG + G G D Sbjct: 284 CYNCQQPGHNSRDCPEERKpreGRNGFTGGSSGFGGGNGGGTGFDSGLTNGFGSGNNGES 343 Query: 219 XXXXXXXXXXXXXXXXXXXXXXXXXXXNNCYNCGQSGHISSYCTQGSRNNGPSGRGCYKC 398 NNC+NC Q GH S+ C + + P R CY C Sbjct: 344 GFGSGGFGGNSNGFGSGGGGQDRGERNNNCFNCQQPGHRSNDCPEPKKEREP--RVCYNC 401 Query: 399 GESGHMARDCVD-------------------------------GNEKQ---VKCYNCNQS 476 + GH +RDC + GN ++ +KC+NC Sbjct: 402 QQPGHNSRDCPEERKpreGRNGFTSGFGGGNDGGFGGGNAEGFGNNEERGPMKCFNCKGE 461 Query: 477 GHMSRECPNGPAEKTCYKCHNTGHIARDC 563 GH S ECP P + C+ C GH + +C Sbjct: 462 GHRSAECPEPP--RGCFNCGEQGHRSNEC 488
Score = 82.8 bits (203), Expect = 6e-16
Identities = 56/190 (29%), Positives = 72/190 (37%), Gaps = 7/190 (3%)
Frame = +3
Query: 24 EQMSQRRLECYNCGQSGHFSRDCTESKRYGSGARSGGCYNCGENGHISRDCPQNXXXXXX 203
+ +R C+NC Q GH S DC E K+ CYNC + GH SRDCP+
Sbjct: 250 QDRGERNNNCFNCQQPGHRSNDCPEPKKE---REPRVCYNCQQPGHNSRDCPEERKpreG 306
Query: 204 XXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXNNCYNCGQSGHISSYCTQ-GSRNNGPSG 380
N G +G S GS NNG SG
Sbjct: 307 RNGFTGGSSGFGGG----------------------NGGGTGFDSGLTNGFGSGNNGESG 344
Query: 381 RGCYKCGESGH---MARDCVDGNEKQVKCYNCNQSGHMSRECPNGPAE---KTCYKCHNT 542
G G + + D E+ C+NC Q GH S +CP E + CY C
Sbjct: 345 FGSGGFGGNSNGFGSGGGGQDRGERNNNCFNCQQPGHRSNDCPEPKKEREPRVCYNCQQP 404
Query: 543 GHIARDCHDD 572
GH +RDC ++
Sbjct: 405 GHNSRDCPEE 414
Score = 53.1 bits (126), Expect = 5e-07
Identities = 26/76 (34%), Positives = 34/76 (44%), Gaps = 3/76 (3%)
Frame = +3
Query: 354 GSRNNGPSGRGCYKCGESGHMARDCVDGNEKQVKCYNCNQSGHMSRECPNGPAE---KTC 524
G N+G SG G D E+ C+NC Q GH S +CP E + C
Sbjct: 225 GGGNSGGSGFGSGGNSNGFGSGGGGQDRGERNNNCFNCQQPGHRSNDCPEPKKEREPRVC 284
Query: 525 YKCHNTGHIARDCHDD 572
Y C GH +RDC ++
Sbjct: 285 YNCQQPGHNSRDCPEE 300
Score = 49.3 bits (116), Expect = 8e-06
Identities = 20/45 (44%), Positives = 26/45 (57%)
Frame = +3
Query: 45 LECYNCGQSGHFSRDCTESKRYGSGARSGGCYNCGENGHISRDCP 179
++C+NC GH S +C E R GC+NCGE GH S +CP
Sbjct: 453 MKCFNCKGEGHRSAECPEPPR--------GCFNCGEQGHRSNECP 489
Score = 31.2 bits (69), Expect = 2.2
Identities = 11/28 (39%), Positives = 15/28 (53%)
Frame = +3
Query: 51 CYNCGQSGHFSRDCTESKRYGSGARSGG 134
C+NCG+ GH S +C + GA G
Sbjct: 475 CFNCGEQGHRSNECPNPAKpreGAEGEG 502
>sp|O76743|GLH4_CAEEL ATP-dependent RNA helicase glh-4 (Germline helicase 4) Length = 1156 Score = 77.8 bits (190), Expect = 2e-14 Identities = 48/174 (27%), Positives = 62/174 (35%), Gaps = 5/174 (2%) Frame = +3 Query: 57 NCGQSGHFSRDCTESK-RYGSGARSGGCYNCGENGHISRDCPQNXXXXXXXXXXXXXXXX 233 N G F + + G R GC+NCGE GHIS++C + Sbjct: 545 NFGNGNTFGEPSDNQRGNWDGGERPRGCHNCGEEGHISKECDKPKVPRFP---------- 594 Query: 234 XXXXXXXXXXXXXXXXXXXXXXNNCYNCGQSGHISSYCTQGSRNNGPSGRGCYKCGESGH 413 C NC Q GH +S C Q GP C CG GH Sbjct: 595 ------------------------CRNCEQLGHFASDCDQPRVPRGP----CRNCGIEGH 626 Query: 414 MARDCVDGNEKQVKCYNCNQSGHMSRECPNGPAE----KTCYKCHNTGHIARDC 563 A DC + C NC Q GH +++C N + C +C GH +C Sbjct: 627 FAVDCDQPKVPRGPCRNCGQEGHFAKDCQNERVRMEPTEPCRRCAEEGHWGYEC 680
Score = 74.3 bits (181), Expect = 2e-13
Identities = 45/163 (27%), Positives = 59/163 (36%), Gaps = 3/163 (1%)
Frame = +3
Query: 36 QRRLECYNCGQSGHFSRDCTESKRYGSGARSGGCYNCGENGHISRDCPQNXXXXXXXXXX 215
+R C+NCG+ GH S++C + K C NC + GH + DC Q
Sbjct: 567 ERPRGCHNCGEEGHISKECDKPK-----VPRFPCRNCEQLGHFASDCDQ----------- 610
Query: 216 XXXXXXXXXXXXXXXXXXXXXXXXXXXXNNCYNCGQSGHISSYCTQGSRNNGPSGRGCYK 395
C NCG GH + C Q GP C
Sbjct: 611 -----------------------PRVPRGPCRNCGIEGHFAVDCDQPKVPRGP----CRN 643
Query: 396 CGESGHMARDCVDGN---EKQVKCYNCNQSGHMSRECPNGPAE 515
CG+ GH A+DC + E C C + GH ECP P +
Sbjct: 644 CGQEGHFAKDCQNERVRMEPTEPCRRCAEEGHWGYECPTRPKD 686
Score = 67.0 bits (162), Expect = 4e-11
Identities = 33/94 (35%), Positives = 43/94 (45%), Gaps = 5/94 (5%)
Frame = +3
Query: 306 CYNCGQSGHISSYCTQGSRNNGPSGRGCYKCGESGHMARDCVDGNEKQVKCYNCNQSGHM 485
C+NCG+ GHIS C + P C C + GH A DC + C NC GH
Sbjct: 572 CHNCGEEGHISKECDKPKVPRFP----CRNCEQLGHFASDCDQPRVPRGPCRNCGIEGHF 627
Query: 486 SREC-----PNGPAEKTCYKCHNTGHIARDCHDD 572
+ +C P GP C C GH A+DC ++
Sbjct: 628 AVDCDQPKVPRGP----CRNCGQEGHFAKDCQNE 657
Score = 49.3 bits (116), Expect = 8e-06
Identities = 27/86 (31%), Positives = 38/86 (44%), Gaps = 1/86 (1%)
Frame = +3
Query: 309 YNCGQSGHISSYCTQGSRNNGPSGRGCYKCGESGHMARDCVDGNEKQVKCYNCNQSGHMS 488
+N G S++ G ++ G G GE R DG E+ C+NC + GH+S
Sbjct: 524 FNTGGGAVKSAFGAAGFGSSSNFGNG-NTFGEPSDNQRGNWDGGERPRGCHNCGEEGHIS 582
Query: 489 RECPNGPAEK-TCYKCHNTGHIARDC 563
+EC + C C GH A DC
Sbjct: 583 KECDKPKVPRFPCRNCEQLGHFASDC 608
Score = 42.0 bits (97), Expect = 0.001
Identities = 19/52 (36%), Positives = 24/52 (46%)
Frame = +3
Query: 24 EQMSQRRLECYNCGQSGHFSRDCTESKRYGSGARSGGCYNCGENGHISRDCP 179
+Q R C NCGQ GHF++DC + C C E GH +CP
Sbjct: 632 DQPKVPRGPCRNCGQEGHFAKDCQNERVRMEPTEP--CRRCAEEGHWGYECP 681
>sp|P34689|GLH1_CAEEL ATP-dependent RNA helicase glh-1 (Germline helicase 1) Length = 763 Score = 70.9 bits (172), Expect = 3e-12 Identities = 38/124 (30%), Positives = 53/124 (42%), Gaps = 36/124 (29%) Frame = +3 Query: 300 NNCYNCGQSGHISSYCTQGSRNNGPSGRGCYKCGESGHMARDCVD--------------- 434 NNC+NC Q GH SS C + + P R CY C + GH +R+C + Sbjct: 158 NNCFNCQQPGHRSSDCPEPRKEREP--RVCYNCQQPGHTSRECTEERKpreGRTGGFGGG 215 Query: 435 -------------------GNEKQ--VKCYNCNQSGHMSRECPNGPAEKTCYKCHNTGHI 551 G E++ +KC+NC GH S ECP P + C+ C GH Sbjct: 216 AGFGNNGGNDGFGGDGGFGGGEERGPMKCFNCKGEGHRSAECPEPP--RGCFNCGEQGHR 273 Query: 552 ARDC 563 + +C Sbjct: 274 SNEC 277
Score = 63.2 bits (152), Expect = 5e-10
Identities = 42/164 (25%), Positives = 55/164 (33%), Gaps = 32/164 (19%)
Frame = +3
Query: 108 YGSGARSGGCYNCGENGHISRDCPQNXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXX 287
+G G R+ C+NC + GH S DCP+
Sbjct: 151 HGGGERNNNCFNCQQPGHRSSDCPEPRKEREPRV-------------------------- 184
Query: 288 XXXXNNCYNCGQSGHISSYCTQGSR--------------------NNGPSGRG------- 386
CYNC Q GH S CT+ + N+G G G
Sbjct: 185 ------CYNCQQPGHTSRECTEERKpreGRTGGFGGGAGFGNNGGNDGFGGDGGFGGGEE 238
Query: 387 -----CYKCGESGHMARDCVDGNEKQVKCYNCNQSGHMSRECPN 503
C+ C GH + +C E C+NC + GH S ECPN
Sbjct: 239 RGPMKCFNCKGEGHRSAECP---EPPRGCFNCGEQGHRSNECPN 279
Score = 63.2 bits (152), Expect = 5e-10
Identities = 36/133 (27%), Positives = 49/133 (36%), Gaps = 2/133 (1%)
Frame = +3
Query: 36 QRRLECYNCGQSGHFSRDCTESKRYGSGARSGGCYNCGENGHISRDCPQNXXXXXXXXXX 215
+R C+NC Q GH S DC E ++ CYNC + GH SR+C +
Sbjct: 155 ERNNNCFNCQQPGHRSSDCPEPRK---EREPRVCYNCQQPGHTSRECTEERKpreGRTGG 211
Query: 216 XXXXXXXXXXXXXXXXXXXXXXXXXXXXN--NCYNCGQSGHISSYCTQGSRNNGPSGRGC 389
C+NC GH S+ C + RGC
Sbjct: 212 FGGGAGFGNNGGNDGFGGDGGFGGGEERGPMKCFNCKGEGHRSAECPE-------PPRGC 264
Query: 390 YKCGESGHMARDC 428
+ CGE GH + +C
Sbjct: 265 FNCGEQGHRSNEC 277
Score = 58.2 bits (139), Expect = 2e-08
Identities = 30/88 (34%), Positives = 38/88 (43%), Gaps = 3/88 (3%)
Frame = +3
Query: 318 GQSGHISSYCTQGSRNNGPSGRGCYKCGESGHMARDCVDGNEKQVKCYNCNQSGHMSREC 497
G SG T G G G GE GH G E+ C+NC Q GH S +C
Sbjct: 120 GGSGGFGGSATGFGSGGGSFGGGNSGFGEGGH------GGGERNNNCFNCQQPGHRSSDC 173
Query: 498 PNGPAE---KTCYKCHNTGHIARDCHDD 572
P E + CY C GH +R+C ++
Sbjct: 174 PEPRKEREPRVCYNCQQPGHTSRECTEE 201
Score = 49.3 bits (116), Expect = 8e-06
Identities = 20/45 (44%), Positives = 26/45 (57%)
Frame = +3
Query: 45 LECYNCGQSGHFSRDCTESKRYGSGARSGGCYNCGENGHISRDCP 179
++C+NC GH S +C E R GC+NCGE GH S +CP
Sbjct: 242 MKCFNCKGEGHRSAECPEPPR--------GCFNCGEQGHRSNECP 278
Score = 29.6 bits (65), Expect = 6.4
Identities = 10/28 (35%), Positives = 14/28 (50%)
Frame = +3
Query: 51 CYNCGQSGHFSRDCTESKRYGSGARSGG 134
C+NCG+ GH S +C + G G
Sbjct: 264 CFNCGEQGHRSNECPNPAKpreGVEGEG 291
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 67,805,915
Number of Sequences: 369166
Number of extensions: 1307558
Number of successful extensions: 8228
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 4178
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7274
length of database: 68,354,980
effective HSP length: 106
effective length of database: 48,773,070
effective search space used: 4877307000
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail