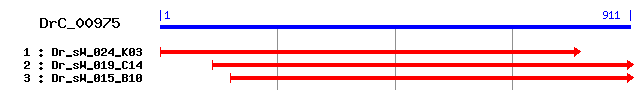
DrC_00975
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00975 (911 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P61980|HNRPK_RAT Heterogeneous nuclear ribonucleoprotein... 81 5e-15 sp|O19049|HNRPK_RABIT Heterogeneous nuclear ribonucleoprote... 80 8e-15 sp|P57722|PCBP3_MOUSE Poly(rC)-binding protein 3 (Alpha-CP3) 70 1e-11 sp|P57721|PCBP3_HUMAN Poly(rC)-binding protein 3 (Alpha-CP3) 70 1e-11 sp|P57723|PCBP4_HUMAN Poly(rC)-binding protein 4 (Alpha-CP4) 61 4e-09 sp|Q61990|PCBP2_MOUSE Poly(rC)-binding protein 2 (Alpha-CP2... 58 4e-08 sp|Q15366|PCBP2_HUMAN Poly(rC)-binding protein 2 (Alpha-CP2... 57 7e-08 sp|Q96I24|FUBP3_HUMAN Far upstream element-binding protein ... 52 2e-06 sp|Q92945|FUBP2_HUMAN Far upstream element-binding protein ... 50 9e-06 sp|P60335|PCBP1_MOUSE Poly(rC)-binding protein 1 (Alpha-CP1... 48 3e-05
>sp|P61980|HNRPK_RAT Heterogeneous nuclear ribonucleoprotein K (dC stretch-binding protein) (CSBP) sp|P61979|HNRPK_MOUSE Heterogeneous nuclear ribonucleoprotein K sp|P61978|HNRPK_HUMAN Heterogeneous nuclear ribonucleoprotein K (hnRNP K) (Transformation up-regulated nuclear protein) (TUNP) Length = 463 Score = 80.9 bits (198), Expect = 5e-15 Identities = 40/105 (38%), Positives = 69/105 (65%) Frame = +1 Query: 55 YIESENEIDLRMVVHESQAGAVIGKGGERIRNLIDSHGMTMIKLYKTFCPNSTDRVVRMV 234 Y S+ + +LR+++H+S AG +IG G +I+ L ++ T IKL++ CP+STDRVV + Sbjct: 138 YKGSDFDCELRLLIHQSLAGGIIGVKGAKIKELRENT-QTTIKLFQECCPHSTDRVVLIG 196 Query: 235 SKMENLMACLENVFELLENQAIKGTRKDYNPDNYDESLAPNYGGF 369 K + ++ C++ + +L+ IKG + Y+P+ YDE+ +YGGF Sbjct: 197 GKPDRVVECIKIILDLISESPIKGRAQPYDPNFYDETY--DYGGF 239
Score = 53.1 bits (126), Expect = 1e-06
Identities = 25/74 (33%), Positives = 45/74 (60%)
Frame = +1
Query: 640 SRVTVPQNLVAAIVGKDGRRLNDLERECGANIEMDIDRMNMGESIFIIRGSPDQIYKAQC 819
++VT+P++L +I+GK G+R+ + E GA+I++D + I I G+ DQI AQ
Sbjct: 390 TQVTIPKDLAGSIIGKGGQRIKQIRHESGASIKIDEPLEGSEDRIITITGTQDQIQNAQY 449
Query: 820 LMQLCARKFADDGF 861
L+Q ++++ F
Sbjct: 450 LLQNSVKQYSGKFF 463
Score = 35.0 bits (79), Expect = 0.29
Identities = 21/92 (22%), Positives = 50/92 (54%), Gaps = 2/92 (2%)
Frame = +1
Query: 19 AKECDYKGSSSKYIESENEIDLRMVVHESQAGAVIGKGGERIRNLIDSHGMTMIKLYKTF 198
A++ + + + + ++ ++LR+++ AGAVIGKGG+ I+ L + ++
Sbjct: 24 AEDMEEEQAFKRSRNTDEMVELRILLQSKNAGAVIGKGGKNIKALRTDYNASV------S 77
Query: 199 CPNST--DRVVRMVSKMENLMACLENVFELLE 288
P+S+ +R++ + + +E + L+ + LE
Sbjct: 78 VPDSSGPERILSISADIETIGEILKKIIPTLE 109
Score = 32.0 bits (71), Expect = 2.5
Identities = 20/72 (27%), Positives = 39/72 (54%)
Frame = +1
Query: 76 IDLRMVVHESQAGAVIGKGGERIRNLIDSHGMTMIKLYKTFCPNSTDRVVRMVSKMENLM 255
I ++ + + AG++IGKGG+RI+ + G + IK+ + S DR++ + +
Sbjct: 388 ITTQVTIPKDLAGSIIGKGGQRIKQIRHESGAS-IKIDEPL-EGSEDRIITITGTQDQ-- 443
Query: 256 ACLENVFELLEN 291
++N LL+N
Sbjct: 444 --IQNAQYLLQN 453
>sp|O19049|HNRPK_RABIT Heterogeneous nuclear ribonucleoprotein K (hnRNP K) Length = 463 Score = 80.1 bits (196), Expect = 8e-15 Identities = 39/102 (38%), Positives = 68/102 (66%) Frame = +1 Query: 64 SENEIDLRMVVHESQAGAVIGKGGERIRNLIDSHGMTMIKLYKTFCPNSTDRVVRMVSKM 243 S+ + +LR+++H+S AG +IG G +I+ L ++ T IKL++ CP+STDRVV + K Sbjct: 141 SDFDCELRLLIHQSLAGGIIGVKGAKIKELRENT-QTTIKLFQECCPHSTDRVVLIGGKP 199 Query: 244 ENLMACLENVFELLENQAIKGTRKDYNPDNYDESLAPNYGGF 369 + ++ C++ + +L+ IKG + Y+P+ YDE+ +YGGF Sbjct: 200 DRVVECIKIILDLISESPIKGRAQPYDPNFYDETY--DYGGF 239
Score = 53.1 bits (126), Expect = 1e-06
Identities = 25/74 (33%), Positives = 45/74 (60%)
Frame = +1
Query: 640 SRVTVPQNLVAAIVGKDGRRLNDLERECGANIEMDIDRMNMGESIFIIRGSPDQIYKAQC 819
++VT+P++L +I+GK G+R+ + E GA+I++D + I I G+ DQI AQ
Sbjct: 390 TQVTIPKDLAGSIIGKGGQRIKQIRHESGASIKIDEPLEGSEDRIITITGTQDQIQNAQY 449
Query: 820 LMQLCARKFADDGF 861
L+Q ++++ F
Sbjct: 450 LLQNSVKQYSGKFF 463
Score = 35.4 bits (80), Expect = 0.22
Identities = 29/125 (23%), Positives = 58/125 (46%), Gaps = 4/125 (3%)
Frame = +1
Query: 19 AKECDYKGSSSKYIESENEIDLRMVVHESQAGAVIGKGGERIRNLIDSHGMTMIKLYKTF 198
A++ + + + + ++ ++LR+++ AGAVIGKGG+ I+ L + ++
Sbjct: 24 AEDMEEEQAFKRSRNTDEMVELRILLQSKNAGAVIGKGGKNIKALRTDYNASV------S 77
Query: 199 CPNST--DRVVRMVSKMENLMACLENVFELLEN--QAIKGTRKDYNPDNYDESLAPNYGG 366
P+S+ +R++ + + +E + L+ + LE Q T P D NY
Sbjct: 78 VPDSSGPERILSISADIETIGEILKKIIPTLEEGLQLPSPTATSQLPLESDAVECLNYQH 137
Query: 367 FISKD 381
F D
Sbjct: 138 FKGSD 142
Score = 32.0 bits (71), Expect = 2.5
Identities = 20/72 (27%), Positives = 39/72 (54%)
Frame = +1
Query: 76 IDLRMVVHESQAGAVIGKGGERIRNLIDSHGMTMIKLYKTFCPNSTDRVVRMVSKMENLM 255
I ++ + + AG++IGKGG+RI+ + G + IK+ + S DR++ + +
Sbjct: 388 ITTQVTIPKDLAGSIIGKGGQRIKQIRHESGAS-IKIDEPL-EGSEDRIITITGTQDQ-- 443
Query: 256 ACLENVFELLEN 291
++N LL+N
Sbjct: 444 --IQNAQYLLQN 453
>sp|P57722|PCBP3_MOUSE Poly(rC)-binding protein 3 (Alpha-CP3) Length = 339 Score = 69.7 bits (169), Expect = 1e-11 Identities = 60/260 (23%), Positives = 105/260 (40%), Gaps = 6/260 (2%) Frame = +1 Query: 64 SENEIDLRMVVHESQAGAVIGKGGERIRNLIDSHGMTMIKLYKTFCPNSTDRVVRMVSKM 243 S+ + LR+VV SQ G++IGKGG +I+ + +S G +++ PNST+R V + Sbjct: 94 SKPPVTLRLVVPASQCGSLIGKGGSKIKEIRESTG-AQVQVAGDMLPNSTERAVTISGTP 152 Query: 244 ENLMACLENVFELLENQAIKGTRKDYNPDNYDESLAPNYGGFISKDSRHPPRRGINNNQS 423 + ++ C++ + ++ KG Y P + G + I + Sbjct: 153 DAIIQCVKQICVVMLESPPKGATIPYRPKPASTPVIFAGGQAYT----------IQGQYA 202 Query: 424 VGHISDLYKSDDYMPHPNPFSRSIPHGYDDGMHNVRPMNNQRNPMLERGGVTDLFGGSIR 603 + H L K PF P Q NP G L Sbjct: 203 IPHPDQLTKLHQLAMQQTPF----------------PPLGQTNPAFP-GEKLPLHSSEEA 245 Query: 604 MNSINDVNSMNMS------RVTVPQNLVAAIVGKDGRRLNDLERECGANIEMDIDRMNMG 765 N + + ++ S +T+P +L+ I+G+ G ++N++ + GA I++ Sbjct: 246 QNLMGQSSGLDASPPASTHELTIPNDLIGCIIGRQGTKINEIRQMSGAQIKIANATEGSS 305 Query: 766 ESIFIIRGSPDQIYKAQCLM 825 E I G+P I AQ L+ Sbjct: 306 ERQITITGTPANISLAQYLI 325
>sp|P57721|PCBP3_HUMAN Poly(rC)-binding protein 3 (Alpha-CP3) Length = 339 Score = 69.7 bits (169), Expect = 1e-11 Identities = 60/260 (23%), Positives = 105/260 (40%), Gaps = 6/260 (2%) Frame = +1 Query: 64 SENEIDLRMVVHESQAGAVIGKGGERIRNLIDSHGMTMIKLYKTFCPNSTDRVVRMVSKM 243 S+ + LR+VV SQ G++IGKGG +I+ + +S G +++ PNST+R V + Sbjct: 94 SKPPVTLRLVVPASQCGSLIGKGGSKIKEIRESTG-AQVQVAGDMLPNSTERAVTISGTP 152 Query: 244 ENLMACLENVFELLENQAIKGTRKDYNPDNYDESLAPNYGGFISKDSRHPPRRGINNNQS 423 + ++ C++ + ++ KG Y P + G + I + Sbjct: 153 DAIIQCVKQICVVMLESPPKGATIPYRPKPASTPVIFAGGQAYT----------IQGQYA 202 Query: 424 VGHISDLYKSDDYMPHPNPFSRSIPHGYDDGMHNVRPMNNQRNPMLERGGVTDLFGGSIR 603 + H L K PF P Q NP G L Sbjct: 203 IPHPDQLTKLHQLAMQQTPF----------------PPLGQTNPAFP-GEKLPLHSSEEA 245 Query: 604 MNSINDVNSMNMS------RVTVPQNLVAAIVGKDGRRLNDLERECGANIEMDIDRMNMG 765 N + + ++ S +T+P +L+ I+G+ G ++N++ + GA I++ Sbjct: 246 QNLMGQSSGLDASPPASTHELTIPNDLIGCIIGRQGTKINEIRQMSGAQIKIANATEGSS 305 Query: 766 ESIFIIRGSPDQIYKAQCLM 825 E I G+P I AQ L+ Sbjct: 306 ERQITITGTPANISLAQYLI 325
>sp|P57723|PCBP4_HUMAN Poly(rC)-binding protein 4 (Alpha-CP4) Length = 403 Score = 61.2 bits (147), Expect = 4e-09 Identities = 54/259 (20%), Positives = 100/259 (38%), Gaps = 2/259 (0%) Frame = +1 Query: 64 SENEIDLRMVVHESQAGAVIGKGGERIRNLIDSHGMTMIKLYKTFCPNSTDRVVRMVSKM 243 S + LR+V+ SQ G++IGK G +I+ + ++ G +++ PNST+R V + Sbjct: 98 SRPPVTLRLVIPASQCGSLIGKAGTKIKEIRETTG-AQVQVAGDLLPNSTERAVTVSGVP 156 Query: 244 ENLMACLENVFELLENQAIKGTRKDYNPD-NYDESLAPNYGGFISKDSRHPPRRGINNNQ 420 + ++ C+ + ++ KG Y+P + L GF + Sbjct: 157 DAIILCVRQICAVILESPPKGATIPYHPSLSLGTVLLSANQGF-----------SVQGQY 205 Query: 421 SVGHISDLYKSDDYMPHPNPFSR-SIPHGYDDGMHNVRPMNNQRNPMLERGGVTDLFGGS 597 +++ K H PF+ S+ G D G Sbjct: 206 GAVTPAEVTKLQQLSSHAVPFATPSVVPGLDPG--------------------------- 238 Query: 598 IRMNSINDVNSMNMSRVTVPQNLVAAIVGKDGRRLNDLERECGANIEMDIDRMNMGESIF 777 + VP +L+ ++G+ G +++++ + GA+I++ GE Sbjct: 239 ---------TQTSSQEFLVPNDLIGCVIGRQGSKISEIRQMSGAHIKIGNQAEGAGERHV 289 Query: 778 IIRGSPDQIYKAQCLMQLC 834 I GSP I AQ L+ C Sbjct: 290 TITGSPVSIALAQYLITAC 308
Score = 30.8 bits (68), Expect = 5.5
Identities = 17/53 (32%), Positives = 28/53 (52%), Gaps = 4/53 (7%)
Frame = +1
Query: 58 IESENEID----LRMVVHESQAGAVIGKGGERIRNLIDSHGMTMIKLYKTFCP 204
+E E E+ LRM++H + G++IGK GE ++ I I + + CP
Sbjct: 8 LEEEPELSITLTLRMLMHGKEVGSIIGKKGETVKR-IREQSSARITISEGSCP 59
>sp|Q61990|PCBP2_MOUSE Poly(rC)-binding protein 2 (Alpha-CP2) (Putative heterogeneous nuclear ribonucleoprotein X) (hnRNP X) (CTBP) (CBP) Length = 362 Score = 57.8 bits (138), Expect = 4e-08 Identities = 60/267 (22%), Positives = 108/267 (40%), Gaps = 11/267 (4%) Frame = +1 Query: 64 SENEIDLRMVVHESQAGAVIGKGGERIRNLIDSHGMTMIKLYKTFCPNSTDRVVRMVSKM 243 S + LR+VV SQ G++IGKGG +I+ + +S G +++ PNST+R + + Sbjct: 94 SRPPVTLRLVVPASQCGSLIGKGGCKIKEIRESTG-AQVQVAGDMLPNSTERAITIAGIP 152 Query: 244 ENLMACLENVFELLENQAIKGTRKDYNPDNYDESLAPNYG------GFISKDSRHPPRRG 405 ++++ C++ + ++ KG Y P + G G S H Sbjct: 153 QSIIECVKQICVVMLESPPKGVTIPYRPKPSSSPVIFAGGQDRYSTGSDSASFPHTTPSM 212 Query: 406 INNNQSVGHISDLYKSDDYMPHPNPFSRSIPHGYDDGMHNVRPMNNQRNPMLE-RGGVTD 582 N G + Y P P +H + M PM G + Sbjct: 213 CLNPDLEGPPLEAYTIQGQYAIPQPDLTK--------LHQL-AMQQSHFPMTHGNTGFSG 263 Query: 583 LFGGSIRMNSI---NDVNSMNMS-RVTVPQNLVAAIVGKDGRRLNDLERECGANIEMDID 750 + S + D ++ S +T+P +L+ I+G+ G ++N++ + GA I++ Sbjct: 264 IESSSPEVKGYWAGLDASAQTTSHELTIPNDLIGCIIGRQGAKINEIRQMSGAQIKIANP 323 Query: 751 RMNMGESIFIIRGSPDQIYKAQCLMQL 831 + I GS I AQ L+ + Sbjct: 324 VEGSTDRQVTITGSAASISLAQYLINV 350
>sp|Q15366|PCBP2_HUMAN Poly(rC)-binding protein 2 (Alpha-CP2) (hnRNP-E2) Length = 365 Score = 57.0 bits (136), Expect = 7e-08 Identities = 62/272 (22%), Positives = 106/272 (38%), Gaps = 16/272 (5%) Frame = +1 Query: 64 SENEIDLRMVVHESQAGAVIGKGGERIRNLIDSHGMTMIKLYKTFCPNSTDRVVRMVSKM 243 S + LR+VV SQ G++IGKGG +I+ + +S G +++ PNST+R + + Sbjct: 94 SRPPVTLRLVVPASQCGSLIGKGGCKIKEIRESTG-AQVQVAGDMLPNSTERAITIAGIP 152 Query: 244 ENLMACLENV----FELLENQAIKGTRKDYNPDNYDESLAPNYG------GFISKDSRHP 393 ++++ C++ + E L KG Y P + G G S H Sbjct: 153 QSIIECVKQICVVMLETLSQSPPKGVTIPYRPKPSSSPVIFAGGQDRYSTGSDSASFPHT 212 Query: 394 PRRGINNNQSVGHISDLYKSDDYMPHPNPFSRSIPHGYDDGMHNVRPMNNQRNPMLERGG 573 N G + Y P P +H + M PM G Sbjct: 213 TPSMCLNPDLEGPPLEAYTIQGQYAIPQPDLTK--------LHQL-AMQQSHFPMTH--G 261 Query: 574 VTDLFGGSIRMNSINDVNSMNMS------RVTVPQNLVAAIVGKDGRRLNDLERECGANI 735 T G + ++ S +T+P +L+ I+G+ G ++N++ + GA I Sbjct: 262 NTGFSGIESSSPEVKGYWGLDASAQTTSHELTIPNDLIGCIIGRQGAKINEIRQMSGAQI 321 Query: 736 EMDIDRMNMGESIFIIRGSPDQIYKAQCLMQL 831 ++ + I GS I AQ L+ + Sbjct: 322 KIANPVEGSTDRQVTITGSAASISLAQYLINV 353
>sp|Q96I24|FUBP3_HUMAN Far upstream element-binding protein 3 (FUSE-binding protein 3) Length = 572 Score = 52.4 bits (124), Expect = 2e-06 Identities = 61/268 (22%), Positives = 106/268 (39%), Gaps = 16/268 (5%) Frame = +1 Query: 70 NEIDLRMVVHE-----SQAGAVIGKGGERIRNLIDSHGMTMIKLYKTFCPNSTDRVVRMV 234 N+ID + E S+ G VIG+GGE I+ L + G+ M+ + P D+ +R+ Sbjct: 156 NDIDSNSTIQEILIPASKVGLVIGRGGETIKQLQERTGVKMVMIQDGPLPTGADKPLRIT 215 Query: 235 SKMENLMACLENVFELL--ENQA-IKGTRKDYNPDNYDESLAPNYGGFISKDSRHPPRRG 405 + E V E++ ++QA +G R D+N S+ + F Sbjct: 216 GDAFKVQQAREMVLEIIREKDQADFRGVRGDFNSRMGGGSIEVSVPRFAVGIVIGRNGEM 275 Query: 406 INNNQSVGHISDLYKSDDYMPHPNPFSRSIPHGYDDGMHNVRPMNNQRNPMLERGGVTDL 585 I Q+ + +K DD + + + D H ++ ER G L Sbjct: 276 IKKIQNDAGVRIQFKPDDGISPER--AAQVMGPPDRCQHAAHIISELILTAQERDGFGGL 333 Query: 586 FGGSIRMN-----SINDVNSMNMSRVTVPQNLVAAIVGKDGRRLNDLERECGANIEMDID 750 R S+ + TVP + ++GK G + + ++ GA++E+ + Sbjct: 334 AAARGRGRGRGDWSVGAPGGVQEITYTVPADKCGLVIGKGGENIKSINQQSGAHVELQRN 393 Query: 751 ---RMNMGESIFIIRGSPDQIYKAQCLM 825 + F IRG P QI A+ L+ Sbjct: 394 PPPNSDPNLRRFTIRGVPQQIEVARQLI 421
Score = 37.7 bits (86), Expect = 0.045
Identities = 25/128 (19%), Positives = 57/128 (44%), Gaps = 20/128 (15%)
Frame = +1
Query: 502 GYDDGMHNVR---------PMNNQRNPMLERGGVTDLFGGSIRMNSINDVNSMNMSRVT- 651
G+ D +H VR P N P+++ ++G ++ ++D + +
Sbjct: 18 GFVDALHRVRQIAAKIDSIPHLNNSTPLVD----PSVYGYGVQKRPLDDGVGNQLGALVH 73
Query: 652 ----------VPQNLVAAIVGKDGRRLNDLERECGANIEMDIDRMNMGESIFIIRGSPDQ 801
VP +V I+G+ G +++ ++ E G I++ + + E ++ G+P+
Sbjct: 74 QRTVITEEFKVPDKMVGFIIGRGGEQISRIQAESGCKIQIASESSGIPERPCVLTGTPES 133
Query: 802 IYKAQCLM 825
I +A+ L+
Sbjct: 134 IEQAKRLL 141
Score = 32.0 bits (71), Expect = 2.5
Identities = 18/73 (24%), Positives = 34/73 (46%), Gaps = 1/73 (1%)
Frame = +1
Query: 646 VTVPQNLVAAIVGKDGRRLNDLERECGANIEMDIDRMNMGESIFIIRGSPDQI-YKAQCL 822
V+VP+ V ++G++G + ++ + G I+ D E + G PD+ + A +
Sbjct: 258 VSVPRFAVGIVIGRNGEMIKKIQNDAGVRIQFKPDDGISPERAAQVMGPPDRCQHAAHII 317
Query: 823 MQLCARKFADDGF 861
+L DGF
Sbjct: 318 SELILTAQERDGF 330
>sp|Q92945|FUBP2_HUMAN Far upstream element-binding protein 2 (FUSE-binding protein 2) (KH type splicing regulatory protein) (KSRP) (p75) Length = 707 Score = 50.1 bits (118), Expect = 9e-06 Identities = 59/260 (22%), Positives = 104/260 (40%), Gaps = 13/260 (5%) Frame = +1 Query: 88 MVVHESQAGAVIGKGGERIRNLIDSHGMTMIKLYKTFCPNSTDRVVRMVSKMENLMACLE 267 +++ +AG VIGKGGE I+ L + G+ MI + + D+ +R++ + E Sbjct: 238 IMIPAGKAGLVIGKGGETIKQLQERAGVKMILIQDGSQNTNVDKPLRIIGDPYKVQQACE 297 Query: 268 NVFELLEN--QAIKGTRKDYNP---DNYDESLAPNYGGFISKDSRHPPRRGINNNQSVGH 432 V ++L N +A G R +Y D + + G + S I Q+ Sbjct: 298 MVMDILRNVTKAGFGDRNEYGSRIGGGIDVPVPRHSVGVVIGRSGEM----IKKIQNDAG 353 Query: 433 ISDLYKSDDYMPHPNPFSRSIPHGYDDGMHNVRPMNNQRNPMLERG-----GVTDLFGGS 597 + +K DD P + G D + + N L G G + GG Sbjct: 354 VRIQFKQDD---GTGPEKIAHIMGPPDRCEHAARIINDLLQSLRSGPPGPPGGPGIPGGR 410 Query: 598 IRMNSINDVNSMNMSRVTVPQNLVAAIVGKDGRRLNDLERECGANIEMDIDRMNMG---E 768 R + ++P + ++G+ G + + ++ GA +E+ G Sbjct: 411 GRGRGQGNWGPGGEMTFSIPTHKCGLVIGRGGENVKAINQQTGAFVEISRQLPPTGTPTS 470 Query: 769 SIFIIRGSPDQIYKAQCLMQ 828 +FIIRGSP QI + L++ Sbjct: 471 KLFIIRGSPQQIDHCRQLIE 490
Score = 40.0 bits (92), Expect = 0.009
Identities = 14/58 (24%), Positives = 34/58 (58%)
Frame = +1
Query: 652 VPQNLVAAIVGKDGRRLNDLERECGANIEMDIDRMNMGESIFIIRGSPDQIYKAQCLM 825
VP +V I+G+ G ++N ++++ G +++ D + E + G+P+ + KA+ ++
Sbjct: 151 VPDGMVGLIIGRGGEQINKIQQDSGCKVQISPDSGGLPERSVSLTGAPESVQKAKMML 208
>sp|P60335|PCBP1_MOUSE Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1) sp|Q15365|PCBP1_HUMAN Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1) (Nucleic acid-binding protein SUB2.3) sp|O19048|PCBP1_RABIT Poly(rC)-binding protein 1 (Alpha-CP1) (hnRNP-E1) Length = 356 Score = 48.1 bits (113), Expect = 3e-05 Identities = 57/273 (20%), Positives = 102/273 (37%), Gaps = 19/273 (6%) Frame = +1 Query: 64 SENEIDLRMVVHESQAGAVIGKGGERIRNLIDSHGMTMIKLYKTFCPNSTDRVVRMVSKM 243 S + LR+VV +Q G++IGKGG +I+ + +S G +++ PNST+R + + Sbjct: 94 SRPPVTLRLVVPATQCGSLIGKGGCKIKEIRESTG-AQVQVAGDMLPNSTERAITIAGVP 152 Query: 244 ENL-----MACLENVFELLENQAIKGTRKDYNPDNYDESLAPNYGGFISKDSRHPPRRGI 408 +++ CL + L ++ + Y P + G D+ P Sbjct: 153 QSVTECVKQICLVMLETLSQSPQGRVMTIPYQPMPASSPVICAGGQDRCSDAAGYPHA-- 210 Query: 409 NNNQSVGHISDLYKSDDYMPHPNPFSRSIPHGYDDGMHNVRPMN--------NQRNPMLE 564 + G D Y G H + P++ Q++ Sbjct: 211 -THDLEGPPLDAYSIQ-------------------GQHTISPLDLAKLNQVARQQSHFAM 250 Query: 565 RGGVTDLFGGSIRMNSIN------DVNSMNMSRVTVPQNLVAAIVGKDGRRLNDLERECG 726 G T G + D ++ +T+P NL+ I+G+ G +N++ + G Sbjct: 251 MHGGTGFAGIDSSSPEVKGYWASLDASTQTTHELTIPNNLIGCIIGRQGANINEIRQMSG 310 Query: 727 ANIEMDIDRMNMGESIFIIRGSPDQIYKAQCLM 825 A I++ I GS I AQ L+ Sbjct: 311 AQIKIANPVEGSSGRQVTITGSAASISLAQYLI 343
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 111,731,227
Number of Sequences: 369166
Number of extensions: 2482320
Number of successful extensions: 6377
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 6027
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6369
length of database: 68,354,980
effective HSP length: 110
effective length of database: 48,034,130
effective search space used: 9270587090
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail