DrC_00924
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
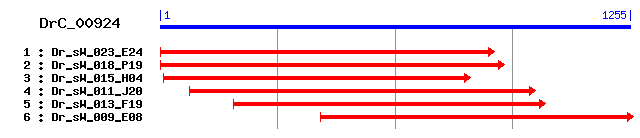
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00924 (1255 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|O62806|MMP13_RABIT Collagenase 3 precursor (Matrix metal... 142 3e-33 sp|P22757|HE_PARLI Hatching enzyme precursor (HE) (HEZ) (En... 141 3e-33 sp|P45452|MMP13_HUMAN Collagenase 3 precursor (Matrix metal... 140 6e-33 sp|Q9Y5R2|MMP24_HUMAN Matrix metalloproteinase-24 precursor... 136 1e-31 sp|Q9R0S2|MMP24_MOUSE Matrix metalloproteinase-24 precursor... 135 2e-31 sp|Q99PW6|MMP24_RAT Matrix metalloproteinase-24 precursor (... 135 2e-31 sp|P91953|HE_HEMPU 50 kDa hatching enzyme precursor (HE) (H... 135 2e-31 sp|P33435|MMP13_MOUSE Collagenase 3 precursor (Matrix metal... 135 2e-31 sp|P23097|MMP13_RAT Collagenase 3 precursor (Matrix metallo... 134 6e-31 sp|O77656|MMP13_BOVIN Collagenase 3 precursor (Matrix metal... 134 7e-31
>sp|O62806|MMP13_RABIT Collagenase 3 precursor (Matrix metalloproteinase-13) (MMP-13) Length = 471 Score = 142 bits (357), Expect = 3e-33 Identities = 117/380 (30%), Positives = 177/380 (46%), Gaps = 21/380 (5%) Frame = +1 Query: 1 ILLSVLFIYSVQSFKSSEEKD--------AEKYLTDFYD--------YDESSGNFEDSIK 132 +LLS +S+ S+E+ D AE YL +Y ++G+ D ++ Sbjct: 10 LLLSWTHCWSLPLLNSNEDDDLSEEDFQFAESYLRSYYHPLNPAGILKKNAAGSMVDRLR 69 Query: 133 ELQKMAGIEQDGELSDEVLSHMNKGRCGGKDREKKNKRYVASGAKWDRGVTYKYYLHLDW 312 E+Q G+E G+L D L+ M + RCG D + N KW + T Y +++ Sbjct: 70 EMQSFFGLEVTGKLDDNTLAIMKQPRCGVPDVGEYN--VFPRTLKWSQ--TNLTYRIVNY 125 Query: 313 MPRTMSEAEVRRAFKVALDYWVRVSNLGFSESRDKVGCKFSISFSGKDHLMYNVDNNFQS 492 P ++ +EV +AFK A W V+ L F+ + ISF K+H Sbjct: 126 TP-DLTHSEVEKAFKKAFKVWSDVTPLNFTRIHNGTA-DIMISFGTKEH----------- 172 Query: 493 QRDPSPLDNVPGGVLAHAYFPR---NGNLHCDNAETWTEGKDSGINLRIVAAHEMGHAIG 663 D P D P G+LAHA+ P G+ H D+ ETWT G NL +VAAHE GH++G Sbjct: 173 -GDFYPFDG-PSGLLAHAFPPGPNYGGDAHFDDDETWTSSS-KGYNLFLVAAHEFGHSLG 229 Query: 664 LHHSQDPSALMAPWYGGYISEAEYRLPNDDTLGAQSIYGARGKRALPSKKRELGKIQSRL 843 L HS+DP ALM P Y Y ++ + LP+DD G QS+YG + P + K Sbjct: 230 LDHSKDPGALMFPIY-TYTGKSHFMLPDDDVQGIQSLYGPGDEDPNPKHPKTPDK----- 283 Query: 844 CNVKSFEDVFTFSDGKVYVLADGGNLFRMNDDKVD-ETFEPVKINEALMN-VDSTFYDNV 1017 C+ D T G+ + D +R++ +VD E F L N +D+ + Sbjct: 284 CDPSLSLDAITSLRGETMIFKD-RFFWRLHPQQVDAELFLTKSFWPELPNRIDAAYEHPA 342 Query: 1018 KKRLIAFKGEKAFSVTGSDI 1077 + + F+G+K ++ G DI Sbjct: 343 RDLIFIFRGKKFWAPNGYDI 362
>sp|P22757|HE_PARLI Hatching enzyme precursor (HE) (HEZ) (Envelysin) (Sea-urchin-hatching proteinase) [Contains: Hatching enzyme 18 kDa form] Length = 587 Score = 141 bits (356), Expect = 3e-33 Identities = 90/232 (38%), Positives = 120/232 (51%), Gaps = 2/232 (0%) Frame = +1 Query: 94 YDESSGNFEDSIKELQKMAGIEQDGELSDEVLSHMNKGRCGGKDREKKNKRYVASGAKWD 273 + E++ N+ +I + Q+ GI Q G L + ++ RCG D +V S W Sbjct: 120 FGEANLNYTSAILDFQEHGGINQTGILDADTAELLSTPRCGVPDVLP----FVTSSITWS 175 Query: 274 RG--VTYKYYLHLDWMPRTMSEAEVRRAFKVALDYWVRVSNLGFSESRDKVGCKFSISFS 447 R VTY + + + + E+RRAF+V W VS L F E D I F Sbjct: 176 RNQPVTYSFGALTSDLNQNDVKDEIRRAFRV----WDDVSGLSFREVPDTTSVDIRIKFG 231 Query: 448 GKDHLMYNVDNNFQSQRDPSPLDNVPGGVLAHAYFPRNGNLHCDNAETWTEGKDSGINLR 627 DH D D GGVLAHA+ PRNG+ H D++ETWTEG SG NL Sbjct: 232 SYDH------------GDGISFDG-RGGVLAHAFLPRNGDAHFDDSETWTEGTRSGTNLF 278 Query: 628 IVAAHEMGHAIGLHHSQDPSALMAPWYGGYISEAEYRLPNDDTLGAQSIYGA 783 VAAHE GH++GL+HS SALM P+Y GY+ +RL NDD G +S+YG+ Sbjct: 279 QVAAHEFGHSLGLYHSTVRSALMYPYYQGYV--PNFRLDNDDIAGIRSLYGS 328
>sp|P45452|MMP13_HUMAN Collagenase 3 precursor (Matrix metalloproteinase-13) (MMP-13) Length = 471 Score = 140 bits (354), Expect = 6e-33 Identities = 113/352 (32%), Positives = 167/352 (47%), Gaps = 14/352 (3%) Frame = +1 Query: 64 AEKYLTDFYD--------YDESSGNFEDSIKELQKMAGIEQDGELSDEVLSHMNKGRCGG 219 AE+YL +Y + ++ + + ++E+Q G+E G+L D L M K RCG Sbjct: 39 AERYLRSYYHPTNLAGILKENAASSMTERLREMQSFFGLEVTGKLDDNTLDVMKKPRCGV 98 Query: 220 KDREKKNKRYVASGAKWDR-GVTYKYYLHLDWMPRTMSEAEVRRAFKVALDYWVRVSNLG 396 D + N KW + +TY+ +++ P M+ +EV +AFK A W V+ L Sbjct: 99 PDVGEYN--VFPRTLKWSKMNLTYRI---VNYTP-DMTHSEVEKAFKKAFKVWSDVTPLN 152 Query: 397 FSESRDKVGCKFSISFSGKDHLMYNVDNNFQSQRDPSPLDNVPGGVLAHAYFPR---NGN 567 F+ D + ISF K+H D P D P G+LAHA+ P G+ Sbjct: 153 FTRLHDGIA-DIMISFGIKEH------------GDFYPFDG-PSGLLAHAFPPGPNYGGD 198 Query: 568 LHCDNAETWTEGKDSGINLRIVAAHEMGHAIGLHHSQDPSALMAPWYGGYISEAEYRLPN 747 H D+ ETWT G NL +VAAHE GH++GL HS+DP ALM P Y Y ++ + LP+ Sbjct: 199 AHFDDDETWTSSS-KGYNLFLVAAHEFGHSLGLDHSKDPGALMFPIY-TYTGKSHFMLPD 256 Query: 748 DDTLGAQSIYGARGKRALPSKKRELGKIQSRLCNVKSFEDVFTFSDGKVYVLADGGNLFR 927 DD G QS+YG + P + K C+ D T G+ + D +R Sbjct: 257 DDVQGIQSLYGPGDEDPNPKHPKTPDK-----CDPSLSLDAITSLRGETMIFKD-RFFWR 310 Query: 928 MNDDKVD-ETFEPVKINEALMNVDSTFYDNVKKRLI-AFKGEKAFSVTGSDI 1077 ++ +VD E F L N Y++ LI F+G K +++ G DI Sbjct: 311 LHPQQVDAELFLTKSFWPELPNRIDAAYEHPSHDLIFIFRGRKFWALNGYDI 362
>sp|Q9Y5R2|MMP24_HUMAN Matrix metalloproteinase-24 precursor (MMP-24) (Membrane-type matrix metalloproteinase 5) (MT-MMP 5) (Membrane-type-5 matrix metalloproteinase) (MT5-MMP) Length = 645 Score = 136 bits (342), Expect = 1e-31 Identities = 114/367 (31%), Positives = 173/367 (47%), Gaps = 51/367 (13%) Frame = +1 Query: 103 SSGNFEDSIKELQKMAGIEQDGELSDEVLSHMNKGRCGGKD-----REKKNKRYVASGAK 267 S+ + ++ +Q+ GI G L + M K RCG D R ++NKRY +G K Sbjct: 103 SAKALQSAVSTMQQFYGIPVTGVLDQTTIEWMKKPRCGVPDHPHLSRRRRNKRYALTGQK 162 Query: 268 W-DRGVTYKYYLHLDWMPRTMSEAEVRRAFKVALDYWVRVSNLGFSE------SRDKVGC 426 W + +TY + ++ P+ + E + R+A + A D W +V+ L F E D+ Sbjct: 163 WRQKHITYSIH---NYTPK-VGELDTRKAIRQAFDVWQKVTPLTFEEVPYHEIKSDRKEA 218 Query: 427 KFSISFSGKDHLMYNVDNNFQSQRDPSPLDNVPGGVLAHAYFPR---NGNLHCDNAETWT 597 I F+ H D SP D GG LAHAYFP G+ H D+ E WT Sbjct: 219 DIMIFFASGFH------------GDSSPFDG-EGGFLAHAYFPGPGIGGDTHFDSDEPWT 265 Query: 598 EGKDS--GINLRIVAAHEMGHAIGLHHSQDPSALMAPWYGGYISEAEYRLPNDDTLGAQS 771 G + G +L +VA HE+GHA+GL HS DPSA+MAP+Y Y+ ++LP DD G Q Sbjct: 266 LGNANHDGNDLFLVAVHELGHALGLEHSSDPSAIMAPFY-QYMETHNFKLPQDDLQGIQK 324 Query: 772 IYGARGK-----RALPS-----------KKRE---------LGKIQS------RLCNVKS 858 IYG + R LP+ +K E LG S +C+ + Sbjct: 325 IYGPPAEPLEPTRPLPTLPVRRIHSPSERKHERQPRPPRPPLGDRPSTPGTKPNICD-GN 383 Query: 859 FEDVFTFSDGKVYVLADGGNLFRMNDDKVDETFEPVKINEALMNVDSTF---YDNVKKRL 1029 F V F G+++V D +R+ +++V E + P++I + + + Y+ R Sbjct: 384 FNTVALFR-GEMFVFKDRW-FWRLRNNRVQEGY-PMQIEQFWKGLPARIDAAYERADGRF 440 Query: 1030 IAFKGEK 1050 + FKG+K Sbjct: 441 VFFKGDK 447
>sp|Q9R0S2|MMP24_MOUSE Matrix metalloproteinase-24 precursor (MMP-24) (Membrane-type matrix metalloproteinase 5) (MT-MMP 5) (Membrane-type-5 matrix metalloproteinase) (MT5-MMP) (MMP-21) Length = 618 Score = 135 bits (341), Expect = 2e-31 Identities = 113/362 (31%), Positives = 171/362 (47%), Gaps = 51/362 (14%) Frame = +1 Query: 118 EDSIKELQKMAGIEQDGELSDEVLSHMNKGRCGGKD-----REKKNKRYVASGAKW-DRG 279 + ++ +Q+ GI G L + M K RCG D R ++NKRY +G KW + Sbjct: 81 QSAVSTMQQFYGIPVTGVLDQTTIEWMKKPRCGVPDHPHLSRRRRNKRYALTGQKWRQKH 140 Query: 280 VTYKYYLHLDWMPRTMSEAEVRRAFKVALDYWVRVSNLGFSE------SRDKVGCKFSIS 441 +TY + ++ P+ + E + R+A + A D W +V+ L F E D+ I Sbjct: 141 ITYSIH---NYTPK-VGELDTRKAIRQAFDVWQKVTPLTFEEVPYHEIKSDRKEADIMIF 196 Query: 442 FSGKDHLMYNVDNNFQSQRDPSPLDNVPGGVLAHAYFPR---NGNLHCDNAETWTEGKDS 612 F+ H D SP D GG LAHAYFP G+ H D+ E WT G + Sbjct: 197 FASGFH------------GDSSPFDG-EGGFLAHAYFPGPGIGGDTHFDSDEPWTLGNAN 243 Query: 613 --GINLRIVAAHEMGHAIGLHHSQDPSALMAPWYGGYISEAEYRLPNDDTLGAQSIYGAR 786 G +L +VA HE+GHA+GL HS DPSA+MAP+Y Y+ ++LP DD G Q IYG Sbjct: 244 HDGNDLFLVAVHELGHALGLEHSNDPSAIMAPFY-QYMETHNFKLPQDDLQGIQKIYGPP 302 Query: 787 GK-----RALPS-----------KKRE---------LGKIQS------RLCNVKSFEDVF 873 + R LP+ +K E LG S +C+ +F V Sbjct: 303 AEPLEPTRPLPTLPVRRIHSPSERKHERHPRPPRPPLGDRPSTPGAKPNICD-GNFNTVA 361 Query: 874 TFSDGKVYVLADGGNLFRMNDDKVDETFEPVKINEALMNVDSTF---YDNVKKRLIAFKG 1044 F G+++V D +R+ +++V E + P++I + + + Y+ R + FKG Sbjct: 362 LFR-GEMFVFKDRW-FWRLRNNRVQEGY-PMQIEQFWKGLPARIDAAYERADGRFVFFKG 418 Query: 1045 EK 1050 +K Sbjct: 419 DK 420
>sp|Q99PW6|MMP24_RAT Matrix metalloproteinase-24 precursor (MMP-24) (Membrane-type matrix metalloproteinase 5) (MT-MMP 5) (Membrane-type-5 matrix metalloproteinase) (MT5-MMP) Length = 618 Score = 135 bits (341), Expect = 2e-31 Identities = 113/362 (31%), Positives = 171/362 (47%), Gaps = 51/362 (14%) Frame = +1 Query: 118 EDSIKELQKMAGIEQDGELSDEVLSHMNKGRCGGKD-----REKKNKRYVASGAKW-DRG 279 + ++ +Q+ GI G L + M K RCG D R ++NKRY +G KW + Sbjct: 81 QSAVSTMQQFYGIPVTGVLDQTTIEWMKKPRCGVPDHPHLSRRRRNKRYALTGQKWRQKH 140 Query: 280 VTYKYYLHLDWMPRTMSEAEVRRAFKVALDYWVRVSNLGFSE------SRDKVGCKFSIS 441 +TY + ++ P+ + E + R+A + A D W +V+ L F E D+ I Sbjct: 141 ITYSIH---NYTPK-VGELDTRKAIRQAFDVWQKVTPLTFEEVPYHEIKSDRKEADIMIF 196 Query: 442 FSGKDHLMYNVDNNFQSQRDPSPLDNVPGGVLAHAYFPR---NGNLHCDNAETWTEGKDS 612 F+ H D SP D GG LAHAYFP G+ H D+ E WT G + Sbjct: 197 FASGFH------------GDSSPFDG-EGGFLAHAYFPGPGIGGDTHFDSDEPWTLGNAN 243 Query: 613 --GINLRIVAAHEMGHAIGLHHSQDPSALMAPWYGGYISEAEYRLPNDDTLGAQSIYGAR 786 G +L +VA HE+GHA+GL HS DPSA+MAP+Y Y+ ++LP DD G Q IYG Sbjct: 244 HDGNDLFLVAVHELGHALGLEHSNDPSAIMAPFY-QYMETHNFKLPQDDLQGIQKIYGPP 302 Query: 787 GK-----RALPS-----------KKRE---------LGKIQS------RLCNVKSFEDVF 873 + R LP+ +K E LG S +C+ +F V Sbjct: 303 AEPLEPTRPLPTLPVRRIHSPSERKHERQPRPPRPPLGDRPSTPGAKPNICD-GNFNTVA 361 Query: 874 TFSDGKVYVLADGGNLFRMNDDKVDETFEPVKINEALMNVDSTF---YDNVKKRLIAFKG 1044 F G+++V D +R+ +++V E + P++I + + + Y+ R + FKG Sbjct: 362 LFR-GEMFVFKDRW-FWRLRNNRVQEGY-PMQIEQFWKGLPARIDAAYERADGRFVFFKG 418 Query: 1045 EK 1050 +K Sbjct: 419 DK 420
>sp|P91953|HE_HEMPU 50 kDa hatching enzyme precursor (HE) (HEZ) (Envelysin) (Sea-urchin-hatching proteinase) [Contains: 38 kDa hatching enzyme; 32 kDa hatching enzyme non-specific; 15 kDa peptide] Length = 591 Score = 135 bits (341), Expect = 2e-31 Identities = 89/231 (38%), Positives = 119/231 (51%), Gaps = 2/231 (0%) Frame = +1 Query: 94 YDESSGNFEDSIKELQKMAGIEQDGELSDEVLSHMNKGRCGGKDREKKNKRYVASGAKWD 273 + E++ N+ +I E Q+ GI Q G L E + ++ RCG D YV G W Sbjct: 123 FGEANINYTSAILEYQQNGGINQTGILDAETAALLDTPRCGVPDILP----YVTGGIAWP 178 Query: 274 RGV--TYKYYLHLDWMPRTMSEAEVRRAFKVALDYWVRVSNLGFSESRDKVGCKFSISFS 447 R V TY + + + +T + E+RRAF+V W VS+L F E D I F Sbjct: 179 RNVAVTYSFGTLSNDLSQTAIKNELRRAFQV----WDDVSSLTFREVVDSSSVDIRIKFG 234 Query: 448 GKDHLMYNVDNNFQSQRDPSPLDNVPGGVLAHAYFPRNGNLHCDNAETWTEGKDSGINLR 627 +H +F Q GGVLAHA+ PRNG+ H D++E WT G +SG NL Sbjct: 235 SYEH---GDGISFDGQ----------GGVLAHAFLPRNGDAHFDDSERWTIGTNSGTNLF 281 Query: 628 IVAAHEMGHAIGLHHSQDPSALMAPWYGGYISEAEYRLPNDDTLGAQSIYG 780 VAAHE GH++GL+HS SALM P+Y GY + L DD G S+YG Sbjct: 282 QVAAHEFGHSLGLYHSDVQSALMYPYYRGY--NPNFNLDRDDIAGITSLYG 330
>sp|P33435|MMP13_MOUSE Collagenase 3 precursor (Matrix metalloproteinase-13) (MMP-13) Length = 472 Score = 135 bits (340), Expect = 2e-31 Identities = 113/351 (32%), Positives = 163/351 (46%), Gaps = 13/351 (3%) Frame = +1 Query: 64 AEKYLTDFYDYDESSGNFE--------DSIKELQKMAGIEQDGELSDEVLSHMNKGRCGG 219 AE YL +Y +G + D ++E+Q G+E G+L D L M K RCG Sbjct: 40 AEHYLKSYYHPATLAGILKKSTVTSTVDRLREMQSFFGLEVTGKLDDPTLDIMRKPRCGV 99 Query: 220 KDREKKNKRYVASGAKWDRGVTYKYYLHLDWMPRTMSEAEVRRAFKVALDYWVRVSNLGF 399 D + N KW + T Y +++ P MS +EV +AF+ A W V+ L F Sbjct: 100 PDVGEYN--VFPRTLKWSQ--TNLTYRIVNYTP-DMSHSEVEKAFRKAFKVWSDVTPLNF 154 Query: 400 SESRDKVGCKFSISFSGKDHLMYNVDNNFQSQRDPSPLDNVPGGVLAHAYFPR---NGNL 570 + D ISF K+H D P D P G+LAHA+ P G+ Sbjct: 155 TRIYDGTA-DIMISFGTKEH------------GDFYPFDG-PSGLLAHAFPPGPNYGGDA 200 Query: 571 HCDNAETWTEGKDSGINLRIVAAHEMGHAIGLHHSQDPSALMAPWYGGYISEAEYRLPND 750 H D+ ETWT G NL IVAAHE+GH++GL HS+DP ALM P Y Y ++ + LP+D Sbjct: 201 HFDDDETWTSSS-KGYNLFIVAAHELGHSLGLDHSKDPGALMFPIY-TYTGKSHFMLPDD 258 Query: 751 DTLGAQSIYGARGKRALPSKKRELGKIQSRLCNVKSFEDVFTFSDGKVYVLADGGNLFRM 930 D G Q +YG + P + K C+ D T G+ + D +R+ Sbjct: 259 DVQGIQFLYGPGDEDPNPKHPKTPEK-----CDPALSLDAITSLRGETMIFKD-RFFWRL 312 Query: 931 NDDKVD-ETFEPVKINEALMN-VDSTFYDNVKKRLIAFKGEKAFSVTGSDI 1077 + +V+ E F L N VD+ + + + F+G K +++ G DI Sbjct: 313 HPQQVEAELFLTKSFWPELPNHVDAAYEHPSRDLMFIFRGRKFWALNGYDI 363
>sp|P23097|MMP13_RAT Collagenase 3 precursor (Matrix metalloproteinase-13) (MMP-13) (UMRCASE) Length = 466 Score = 134 bits (337), Expect = 6e-31 Identities = 112/351 (31%), Positives = 163/351 (46%), Gaps = 13/351 (3%) Frame = +1 Query: 64 AEKYLTDFYDYDESSGNFE--------DSIKELQKMAGIEQDGELSDEVLSHMNKGRCGG 219 AE YL +Y +G + D ++E+Q G++ G+L D L M K RCG Sbjct: 34 AEHYLKSYYHPVTLAGILKKSTVTSTVDRLREMQSFFGLDVTGKLDDPTLDIMRKPRCGV 93 Query: 220 KDREKKNKRYVASGAKWDRGVTYKYYLHLDWMPRTMSEAEVRRAFKVALDYWVRVSNLGF 399 D N KW + T Y +++ P +S +EV +AF+ A W V+ L F Sbjct: 94 PDVGVYN--VFPRTLKWSQ--TNLTYRIVNYTP-DISHSEVEKAFRKAFKVWSDVTPLNF 148 Query: 400 SESRDKVGCKFSISFSGKDHLMYNVDNNFQSQRDPSPLDNVPGGVLAHAYFPR---NGNL 570 + D ISF K+H D P D P G+LAHA+ P G+ Sbjct: 149 TRIHDGTA-DIMISFGTKEH------------GDFYPFDG-PSGLLAHAFPPGPNLGGDA 194 Query: 571 HCDNAETWTEGKDSGINLRIVAAHEMGHAIGLHHSQDPSALMAPWYGGYISEAEYRLPND 750 H D+ ETWT G NL IVAAHE+GH++GL HS+DP ALM P Y Y ++ + LP+D Sbjct: 195 HFDDDETWTSSS-KGYNLFIVAAHELGHSLGLDHSKDPGALMFPIY-TYTGKSHFMLPDD 252 Query: 751 DTLGAQSIYGARGKRALPSKKRELGKIQSRLCNVKSFEDVFTFSDGKVYVLADGGNLFRM 930 D G QS+YG + P + K C+ D T G+ + D +R+ Sbjct: 253 DVQGIQSLYGPGDEDPNPKHPKTPEK-----CDPALSLDAITSLRGETMIFKD-RFFWRL 306 Query: 931 NDDKVD-ETFEPVKINEALMN-VDSTFYDNVKKRLIAFKGEKAFSVTGSDI 1077 + +V+ E F L N VD+ + + + F+G K +++ G DI Sbjct: 307 HPQQVEPELFLTKSFWPELPNHVDAAYEHPSRDLMFIFRGRKFWALNGYDI 357
>sp|O77656|MMP13_BOVIN Collagenase 3 precursor (Matrix metalloproteinase-13) (MMP-13) Length = 471 Score = 134 bits (336), Expect = 7e-31 Identities = 114/368 (30%), Positives = 168/368 (45%), Gaps = 17/368 (4%) Frame = +1 Query: 64 AEKYLTDFYDYDESSGNFE--------DSIKELQKMAGIEQDGELSDEVLSHMNKGRCGG 219 AE YL +Y +G + D ++E+Q G+E G L D L M K RCG Sbjct: 39 AESYLKSYYYPQNPAGILKKTAASSVIDRLREMQSFFGLEVTGRLDDNTLDIMKKPRCGV 98 Query: 220 KDREKKNKRYVASGAKWDR-GVTYKYYLHLDWMPRTMSEAEVRRAFKVALDYWVRVSNLG 396 D + N KW + +TY+ +++ P ++ +EV +AF+ A W V+ L Sbjct: 99 PDVGEYN--VFPRTLKWSKMNLTYRI---VNYTP-DLTHSEVEKAFRKAFKVWSDVTPLN 152 Query: 397 FSESRDKVGCKFSISFSGKDHLMYNVDNNFQSQRDPSPLDNVPGGVLAHAYFPR---NGN 567 F+ + ISF K+H D P D P G+LAHA+ P G+ Sbjct: 153 FTRIHNGTA-DIMISFGTKEH------------GDFYPFDG-PSGLLAHAFPPGPNYGGD 198 Query: 568 LHCDNAETWTEGKDSGINLRIVAAHEMGHAIGLHHSQDPSALMAPWYGGYISEAEYRLPN 747 H D+ ETWT G NL +VAAHE GH++GL HS+DP ALM P Y Y ++ + LP+ Sbjct: 199 AHFDDDETWTSSS-KGYNLFLVAAHEFGHSLGLDHSKDPGALMFPIY-TYTGKSHFMLPD 256 Query: 748 DDTLGAQSIYGARGKRALPSKKRELGKIQSRLCNVKSFEDVFTFSDGKVYVLADGGNLFR 927 DD G QS+YG + + K C+ D T G+ + D +R Sbjct: 257 DDVQGIQSLYGPGDEDPYSKHPKTPDK-----CDPSLSLDAITSLRGETLIFKD-RFFWR 310 Query: 928 MNDDKVD-ETFEPVKINEALMNVDSTFYDNVKKRLI-AFKGEKAFSVTGSDIEN---LKV 1092 ++ +V+ E F L N Y++ LI F+G K ++++G DI K+ Sbjct: 311 LHPQQVEAELFLTKSFGPELPNRIDAAYEHPSHDLIFIFRGRKFWALSGYDILEDYPKKI 370 Query: 1093 EELVISDH 1116 EL H Sbjct: 371 SELGFPKH 378
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.317 0.135 0.399
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 148,694,957
Number of Sequences: 369166
Number of extensions: 3248828
Number of successful extensions: 10149
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 9396
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 9930
length of database: 68,354,980
effective HSP length: 113
effective length of database: 47,479,925
effective search space used: 14433897200
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
Cluster detail