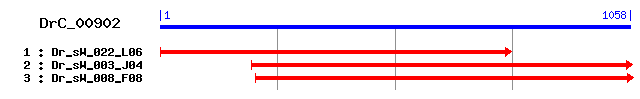
DrC_00902
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00902 (1058 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q9UBX3|DIC_HUMAN Mitochondrial dicarboxylate carrier 334 2e-91 sp|Q9QZD8|DIC_MOUSE Mitochondrial dicarboxylate carrier 324 3e-88 sp|Q9CR62|M2OM_MOUSE Mitochondrial 2-oxoglutarate/malate ca... 183 6e-46 sp|Q02978|M2OM_HUMAN Mitochondrial 2-oxoglutarate/malate ca... 181 2e-45 sp|P22292|M2OM_BOVIN Mitochondrial 2-oxoglutarate/malate ca... 181 3e-45 sp|P97700|M2OM_RAT Mitochondrial 2-oxoglutarate/malate carr... 176 1e-43 sp|P56499|UCP3_RAT Mitochondrial uncoupling protein 3 (UCP 3) 162 1e-39 sp|P56501|UCP3_MOUSE Mitochondrial uncoupling protein 3 (UC... 161 3e-39 sp|O97562|UCP2_PIG Mitochondrial uncoupling protein 2 (UCP 2) 161 3e-39 sp|O77792|UCP3_BOVIN Mitochondrial uncoupling protein 3 (UC... 160 4e-39
>sp|Q9UBX3|DIC_HUMAN Mitochondrial dicarboxylate carrier Length = 287 Score = 334 bits (857), Expect = 2e-91 Identities = 167/278 (60%), Positives = 208/278 (74%), Gaps = 4/278 (1%) Frame = +3 Query: 57 KIGSWYFGGIASAMAAACTHPLDLLKVHLQTQQKKEFGMAGMAIKVVKTDGILALYNGLS 236 ++ WYFGG+AS AA CTHPLDLLKVHLQTQQ+ + M GMA++VV+TDGILALY+GLS Sbjct: 6 RVSRWYFGGLASCGAACCTHPLDLLKVHLQTQQEVKLRMTGMALRVVRTDGILALYSGLS 65 Query: 237 ASLLRQLTYSMTRFGLYETYK----ASKKGATPFYEKAGVSMVSGFIGGIVGNPADMVNV 404 ASL RQ+TYS+TRF +YET + +G PF+EK + VSG GG VG PAD+VNV Sbjct: 66 ASLCRQMTYSLTRFAIYETVRDRVAKGSQGPLPFHEKVLLGSVSGLAGGFVGTPADLVNV 125 Query: 405 RMQNDMKVAKELRRNYKNAVDGMFRVAREEGVVVLFSGVSMTATRAAFMTFGQLAFYDLF 584 RMQND+K+ + RRNY +A+DG++RVAREEG+ LFSG +M ++R A +T GQL+ YD Sbjct: 126 RMQNDVKLPQGQRRNYAHALDGLYRVAREEGLRRLFSGATMASSRGALVTVGQLSCYDQA 185 Query: 585 KQMMIQSGLFSDNIITHFTASTGAAGIATLMTQPFDVMKTRLMNAKPGTYSGLLECGADI 764 KQ+++ +G SDNI THF AS A G AT + QP DV+KTRLMN+K G Y G+ C + Sbjct: 186 KQLVLSTGYLSDNIFTHFVASFIAGGCATFLCQPLDVLKTRLMNSK-GEYQGVFHCAVET 244 Query: 765 MKTGPLGFFKGLVPAFIRLGPQTILTFVFLEQLRMNFG 878 K GPL F+KGLVPA IRL P T+LTFVFLEQLR NFG Sbjct: 245 AKLGPLAFYKGLVPAGIRLIPHTVLTFVFLEQLRKNFG 282
>sp|Q9QZD8|DIC_MOUSE Mitochondrial dicarboxylate carrier Length = 287 Score = 324 bits (830), Expect = 3e-88 Identities = 162/274 (59%), Positives = 200/274 (72%), Gaps = 4/274 (1%) Frame = +3 Query: 69 WYFGGIASAMAAACTHPLDLLKVHLQTQQKKEFGMAGMAIKVVKTDGILALYNGLSASLL 248 WYFGG+AS AA CTHPLDLLKVHLQTQQ+ + M GMA++VV+TDG LALYNGLSASL Sbjct: 9 WYFGGLASCGAACCTHPLDLLKVHLQTQQEVKLRMTGMALQVVRTDGFLALYNGLSASLC 68 Query: 249 RQLTYSMTRFGLYETYK----ASKKGATPFYEKAGVSMVSGFIGGIVGNPADMVNVRMQN 416 RQ+TYS+TRF +YET + +G PFY K + +SG GG VG PAD+VNVRMQN Sbjct: 69 RQMTYSLTRFAIYETMRDYMTKDSQGPLPFYNKVLLGGISGLTGGFVGTPADLVNVRMQN 128 Query: 417 DMKVAKELRRNYKNAVDGMFRVAREEGVVVLFSGVSMTATRAAFMTFGQLAFYDLFKQMM 596 DMK+ RRNY +A+DG++RVAREE + LFSG +M ++R A +T GQL+ YD KQ++ Sbjct: 129 DMKLPPSQRRNYSHALDGLYRVAREESLRKLFSGATMASSRGALVTVGQLSCYDQAKQLV 188 Query: 597 IQSGLFSDNIITHFTASTGAAGIATLMTQPFDVMKTRLMNAKPGTYSGLLECGADIMKTG 776 + +G SDNI THF +S A G AT + QP DV+KTRLMN+K G Y G+ C + K G Sbjct: 189 LSTGYLSDNIFTHFVSSFIAGGCATFLCQPLDVLKTRLMNSK-GEYQGVFHCAMETAKLG 247 Query: 777 PLGFFKGLVPAFIRLGPQTILTFVFLEQLRMNFG 878 P FFKGL PA IRL P T+LTF+FLEQLR +FG Sbjct: 248 PQAFFKGLFPAGIRLIPHTVLTFMFLEQLRKHFG 281
>sp|Q9CR62|M2OM_MOUSE Mitochondrial 2-oxoglutarate/malate carrier protein (OGCP) (Solute carrier family 25 member 11) Length = 314 Score = 183 bits (465), Expect = 6e-46 Identities = 111/287 (38%), Positives = 156/287 (54%), Gaps = 14/287 (4%) Frame = +3 Query: 45 RPQDKIGSWYFGGIASAMAAACTHPLDLLKVHLQTQ----QKKEFGMAGMAI-KVVKTDG 209 R K + FGG+A A PLDL+K +Q + +E+ + A+ ++KT+G Sbjct: 17 RTSPKSVKFLFGGLAGMGATVFVQPLDLVKNRMQLSGEGAKTREYKTSFHALTSILKTEG 76 Query: 210 ILALYNGLSASLLRQLTYSMTRFGLYETYKASKKGA--TP--FYEKAGVSMVSGFIGGIV 377 + +Y GLSA LLRQ TY+ TR G+Y GA TP F KA + M +G G V Sbjct: 77 LKGIYTGLSAGLLRQATYTTTRLGIYTVLFERLTGADGTPPGFLLKALIGMTAGATGAFV 136 Query: 378 GNPADMVNVRMQNDMKVAKELRRNYKNAVDGMFRVAREEGVVVLFSGVSMTATRAAFMTF 557 G PA++ +RM D ++ + RR YKN + + R+AREEGV L+ G T RA + Sbjct: 137 GTPAEVALIRMTADGRLPADQRRGYKNVFNALVRIAREEGVPTLWRGCIPTMARAVVVNA 196 Query: 558 GQLAFYDLFKQMMIQSGLFSDNIITHFTASTGAAGIATLMTQPFDVMKTRLMN-----AK 722 QLA Y KQ ++ SG FSDNI+ HF AS + + T + P D++KTR+ N K Sbjct: 197 AQLASYSQSKQFLLDSGYFSDNILCHFCASMISGLVTTAASMPVDIVKTRIQNMRMIDGK 256 Query: 723 PGTYSGLLECGADIMKTGPLGFFKGLVPAFIRLGPQTILTFVFLEQL 863 P +GL + G +KG P + RLGP T+LTF+FLEQ+ Sbjct: 257 PEYKNGLDVLLKVVRYEGFFSLWKGFTPYYARLGPHTVLTFIFLEQM 303
>sp|Q02978|M2OM_HUMAN Mitochondrial 2-oxoglutarate/malate carrier protein (OGCP) (Solute carrier family 25 member 11) Length = 314 Score = 181 bits (460), Expect = 2e-45 Identities = 109/287 (37%), Positives = 154/287 (53%), Gaps = 14/287 (4%) Frame = +3 Query: 45 RPQDKIGSWYFGGIASAMAAACTHPLDLLKVHLQTQ----QKKEFGMAGMAI-KVVKTDG 209 R K + FGG+A A PLDL+K +Q + +E+ + A+ ++K +G Sbjct: 17 RTSPKSVKFLFGGLAGMGATVFVQPLDLVKNRMQLSGEGAKTREYKTSFHALTSILKAEG 76 Query: 210 ILALYNGLSASLLRQLTYSMTRFGLYETYKASKKGA--TP--FYEKAGVSMVSGFIGGIV 377 + +Y GLSA LLRQ TY+ TR G+Y GA TP F KA + M +G G V Sbjct: 77 LRGIYTGLSAGLLRQATYTTTRLGIYTVLFERLTGADGTPPGFLLKAVIGMTAGATGAFV 136 Query: 378 GNPADMVNVRMQNDMKVAKELRRNYKNAVDGMFRVAREEGVVVLFSGVSMTATRAAFMTF 557 G PA++ +RM D ++ + RR YKN + + R+ REEGV+ L+ G T RA + Sbjct: 137 GTPAEVALIRMTADGRLPADQRRGYKNVFNALIRITREEGVLTLWRGCIPTMARAVVVNA 196 Query: 558 GQLAFYDLFKQMMIQSGLFSDNIITHFTASTGAAGIATLMTQPFDVMKTRLMN-----AK 722 QLA Y KQ ++ SG FSDNI+ HF AS + + T + P D+ KTR+ N K Sbjct: 197 AQLASYSQSKQFLLDSGYFSDNILCHFCASMISGLVTTAASMPVDIAKTRIQNMRMIDGK 256 Query: 723 PGTYSGLLECGADIMKTGPLGFFKGLVPAFIRLGPQTILTFVFLEQL 863 P +GL + G +KG P + RLGP T+LTF+FLEQ+ Sbjct: 257 PEYKNGLDVLFKVVRYEGFFSLWKGFTPYYARLGPHTVLTFIFLEQM 303
>sp|P22292|M2OM_BOVIN Mitochondrial 2-oxoglutarate/malate carrier protein (OGCP) Length = 314 Score = 181 bits (459), Expect = 3e-45 Identities = 109/287 (37%), Positives = 155/287 (54%), Gaps = 14/287 (4%) Frame = +3 Query: 45 RPQDKIGSWYFGGIASAMAAACTHPLDLLKVHLQTQ----QKKEFGMAGMA-IKVVKTDG 209 R K + FGG+A A PLDL+K +Q + +E+ + A I +++ +G Sbjct: 17 RTSPKSVKFLFGGLAGMGATVFVQPLDLVKNRMQLSGEGAKTREYKTSFHALISILRAEG 76 Query: 210 ILALYNGLSASLLRQLTYSMTRFGLYETYKASKKGA--TP--FYEKAGVSMVSGFIGGIV 377 + +Y GLSA LLRQ TY+ TR G+Y GA TP F KA + M +G G V Sbjct: 77 LRGIYTGLSAGLLRQATYTTTRLGIYTVLFERLTGADGTPPGFLLKAVIGMTAGATGAFV 136 Query: 378 GNPADMVNVRMQNDMKVAKELRRNYKNAVDGMFRVAREEGVVVLFSGVSMTATRAAFMTF 557 G PA++ +RM D ++ + RR YKN + +FR+ +EEGV L+ G T RA + Sbjct: 137 GTPAEVALIRMTADGRLPVDQRRGYKNVFNALFRIVQEEGVPTLWRGCIPTMARAVVVNA 196 Query: 558 GQLAFYDLFKQMMIQSGLFSDNIITHFTASTGAAGIATLMTQPFDVMKTRLMN-----AK 722 QLA Y KQ ++ SG FSDNI+ HF AS + + T + P D++KTR+ N K Sbjct: 197 AQLASYSQSKQFLLDSGYFSDNILCHFCASMISGLVTTAASMPVDIVKTRIQNMRMIDGK 256 Query: 723 PGTYSGLLECGADIMKTGPLGFFKGLVPAFIRLGPQTILTFVFLEQL 863 P +GL + G +KG P + RLGP T+LTF+FLEQ+ Sbjct: 257 PEYKNGLDVLVKVVRYEGFFSLWKGFTPYYARLGPHTVLTFIFLEQM 303
>sp|P97700|M2OM_RAT Mitochondrial 2-oxoglutarate/malate carrier protein (OGCP) (Solute carrier family 25 member 11) Length = 314 Score = 176 bits (446), Expect = 1e-43 Identities = 108/287 (37%), Positives = 153/287 (53%), Gaps = 14/287 (4%) Frame = +3 Query: 45 RPQDKIGSWYFGGIASAMAAACTHPLDLLKVHLQTQ----QKKEFGMAGMAI-KVVKTDG 209 R K + FGG+A A PLDL+ +Q + +E+ + A+ ++K +G Sbjct: 17 RTSPKSVKFLFGGLAGMGATVFVQPLDLVXNRMQLSGEGAKTREYKTSFHALTSILKAEG 76 Query: 210 ILALYNGLSASLLRQLTYSMTRFGLYETYKASKKGA--TP--FYEKAGVSMVSGFIGGIV 377 + +Y GLSA LLRQ TY+ TR G+Y GA TP F KA + M +G G V Sbjct: 77 LRGIYTGLSAGLLRQATYTTTRLGIYTVLFERLTGADGTPPGFLLKALIGMTAGATGAFV 136 Query: 378 GNPADMVNVRMQNDMKVAKELRRNYKNAVDGMFRVAREEGVVVLFSGVSMTATRAAFMTF 557 G PA++ +RM D ++ + RR YKN + + R+AREEGV L+ G T RA + Sbjct: 137 GPPAEVALIRMTADGRLPADQRRGYKNVFNALIRIAREEGVPTLWRGCIPTMARAVVVNA 196 Query: 558 GQLAFYDLFKQMMIQSGLFSDNIITHFTASTGAAGIATLMTQPFDVMKTRLMN-----AK 722 QLA Y KQ ++ SG FSDNI+ HF A + + T + P D++KTR+ N K Sbjct: 197 AQLASYSQSKQFLLDSGYFSDNILCHFCAIMISGLVTTAASMPVDIVKTRIQNMRMIDEK 256 Query: 723 PGTYSGLLECGADIMKTGPLGFFKGLVPAFIRLGPQTILTFVFLEQL 863 P +GL + G +KG P + RLGP T+LTF+FLEQ+ Sbjct: 257 PEYKNGLDVLLKVVRYEGFFSLWKGFTPYYARLGPHTVLTFIFLEQM 303
>sp|P56499|UCP3_RAT Mitochondrial uncoupling protein 3 (UCP 3) Length = 308 Score = 162 bits (410), Expect = 1e-39 Identities = 96/285 (33%), Positives = 150/285 (52%), Gaps = 12/285 (4%) Frame = +3 Query: 48 PQDKIGSWYFGGIASAMAAACTHPLDLLKVHLQTQ------QKKEF-GMAGMAIKVVKTD 206 P + + G A+ A T PLD KV LQ Q Q ++ G+ G + +V+T+ Sbjct: 10 PPTTVVKFLGAGTAACFADLLTFPLDTAKVRLQIQGENPGVQSVQYRGVLGTILTMVRTE 69 Query: 207 GILALYNGLSASLLRQLTYSMTRFGLYETYKASKKGATPFYEKAGVSMVSGFIGGIVG-- 380 G + Y+GL A L RQ++++ R GLY++ K + + +++G G + Sbjct: 70 GPRSPYSGLVAGLHRQMSFASIRIGLYDSVKQFYTPKGTDHSSVAIRILAGCTTGAMAVT 129 Query: 381 --NPADMVNVRMQNDMKVAKELRRNYKNAVDGMFRVAREEGVVVLFSGVSMTATRAAFMT 554 P D+V VR Q +++ R Y+ +D +AREEGV L+ G TR A + Sbjct: 130 CAQPTDVVKVRFQAMIRLGTGGERKYRGTMDAYRTIAREEGVRGLWKGTWPNITRNAIVN 189 Query: 555 FGQLAFYDLFKQMMIQSGLFSDNIITHFTASTGAAGIATLMTQPFDVMKTRLMNAKPGTY 734 ++ YD+ K+ ++ S LF+DN HF ++ GA AT++ P DV+KTR MNA PG Y Sbjct: 190 CAEMVTYDIIKEKLLDSHLFTDNFPCHFVSAFGAGFCATVVASPVDVVKTRYMNAPPGRY 249 Query: 735 SGLLECGAD-IMKTGPLGFFKGLVPAFIRLGPQTILTFVFLEQLR 866 L C + + GP F+KG +P+F+RLG ++ FV EQL+ Sbjct: 250 RSPLHCMLRMVAQEGPTAFYKGFMPSFLRLGSWNVMMFVTYEQLK 294
>sp|P56501|UCP3_MOUSE Mitochondrial uncoupling protein 3 (UCP 3) Length = 308 Score = 161 bits (408), Expect = 3e-39 Identities = 99/287 (34%), Positives = 154/287 (53%), Gaps = 14/287 (4%) Frame = +3 Query: 48 PQDKIGSWYFGGIASAMAAACTHPLDLLKVHLQTQ------QKKEF-GMAGMAIKVVKTD 206 P + + G A+ A T PLD KV LQ Q Q ++ G+ G + +V+T+ Sbjct: 10 PPTTVVKFLGAGTAACFADLLTFPLDTAKVRLQIQGENPGAQSVQYRGVLGTILTMVRTE 69 Query: 207 GILALYNGLSASLLRQLTYSMTRFGLYETYKA--SKKGATPFYEKAGVSMVSGFIGGIVG 380 G + Y+GL A L RQ++++ R GLY++ K + KGA + + +++G G + Sbjct: 70 GPRSPYSGLVAGLHRQMSFASIRIGLYDSVKQFYTPKGAD--HSSVAIRILAGCTTGAMA 127 Query: 381 ----NPADMVNVRMQNDMKVAKELRRNYKNAVDGMFRVAREEGVVVLFSGVSMTATRAAF 548 P D+V VR Q +++ R Y+ +D +AREEGV L+ G TR A Sbjct: 128 VTCAQPTDVVKVRFQAMIRLGTGGERKYRGTMDAYRTIAREEGVRGLWKGTWPNITRNAI 187 Query: 549 MTFGQLAFYDLFKQMMIQSGLFSDNIITHFTASTGAAGIATLMTQPFDVMKTRLMNAKPG 728 + ++ YD+ K+ +++S LF+DN HF ++ GA AT++ P DV+KTR MNA G Sbjct: 188 VNCAEMVTYDIIKEKLLESHLFTDNFPCHFVSAFGAGFCATVVASPVDVVKTRYMNAPLG 247 Query: 729 TYSGLLECGAD-IMKTGPLGFFKGLVPAFIRLGPQTILTFVFLEQLR 866 Y L C + + GP F+KG VP+F+RLG ++ FV EQL+ Sbjct: 248 RYRSPLHCMLKMVAQEGPTAFYKGFVPSFLRLGAWNVMMFVTYEQLK 294
>sp|O97562|UCP2_PIG Mitochondrial uncoupling protein 2 (UCP 2) Length = 309 Score = 161 bits (407), Expect = 3e-39 Identities = 99/281 (35%), Positives = 149/281 (53%), Gaps = 19/281 (6%) Frame = +3 Query: 81 GIASAMAAACTHPLDLLKVHLQTQQKKEF-----------GMAGMAIKVVKTDGILALYN 227 G A+ +A T PLD KV LQ Q ++ G+ G + +V+ +G +LYN Sbjct: 21 GTAACIADLITFPLDTAKVRLQIQGERRGPVQAAASAQYRGVLGTILTMVRNEGPRSLYN 80 Query: 228 GLSASLLRQLTYSMTRFGLYETYKAS-KKGATPFYEKAGVSM------VSGFIGGIVGNP 386 GL A L RQ++++ R GLY++ K KG+ E AG+ +G + V P Sbjct: 81 GLVAGLQRQMSFASVRIGLYDSVKHFYTKGS----EHAGIGSRLLAGSTTGALAVAVAQP 136 Query: 387 ADMVNVRMQNDMKVAKELRRNYKNAVDGMFRVAREEGVVVLFSGVSMTATRAAFMTFGQL 566 D+V VR Q + R Y++ VD +AREEG+ L+ G S R A + +L Sbjct: 137 TDVVKVRFQAQARAGGG--RRYRSTVDAYKTIAREEGLRGLWKGTSPNVARNAIVNCAEL 194 Query: 567 AFYDLFKQMMIQSGLFSDNIITHFTASTGAAGIATLMTQPFDVMKTRLMNAKPGTYSGLL 746 YDL K ++++ L +D++ HFT++ GA T++ P DV+KTR MN+ PG YS Sbjct: 195 VTYDLIKDTLLKADLMTDDLPCHFTSAFGAGFCTTVIASPVDVVKTRYMNSAPGQYSSAG 254 Query: 747 ECGADIM-KTGPLGFFKGLVPAFIRLGPQTILTFVFLEQLR 866 C ++ K GP F+KG P+F+RLG ++ FV EQL+ Sbjct: 255 HCALTMLQKEGPRAFYKGFTPSFLRLGSWNVVMFVTYEQLK 295
>sp|O77792|UCP3_BOVIN Mitochondrial uncoupling protein 3 (UCP 3) Length = 311 Score = 160 bits (406), Expect = 4e-39 Identities = 95/289 (32%), Positives = 147/289 (50%), Gaps = 15/289 (5%) Frame = +3 Query: 45 RPQDKIGSWYFGGIASAMAAACTHPLDLLKVHLQTQQKKEFGMA----------GMAIKV 194 RP + G A+ A T PLD KV LQ Q + + +A G + + Sbjct: 9 RPPTTSVKFLAAGTAACFADLLTFPLDTAKVRLQIQGENQAALAARSAQYRGVLGTILTM 68 Query: 195 VKTDGILALYNGLSASLLRQLTYSMTRFGLYETYKA--SKKGA--TPFYEKAGVSMVSGF 362 V+T+G +LY+GL A L RQ++++ R GLY++ K + KG+ + + +G Sbjct: 69 VRTEGPRSLYSGLVAGLQRQMSFASIRIGLYDSVKQFYTPKGSDHSSIITRILAGCTTGA 128 Query: 363 IGGIVGNPADMVNVRMQNDMKVAKELRRNYKNAVDGMFRVAREEGVVVLFSGVSMTATRA 542 + P D+V +R Q M R Y +D +AREEGV L+ G+ TR Sbjct: 129 MAVTCAQPTDVVKIRFQASMHTGLGGNRKYSGTMDAYRTIAREEGVRGLWKGILPNITRN 188 Query: 543 AFMTFGQLAFYDLFKQMMIQSGLFSDNIITHFTASTGAAGIATLMTQPFDVMKTRLMNAK 722 A + G++ YD+ K+ ++ L +DN HF ++ GA ATL+ P DV+KTR MN+ Sbjct: 189 AIVNCGEMVTYDIIKEKLLDYHLLTDNFPCHFVSAFGAGFCATLVASPVDVVKTRYMNSP 248 Query: 723 PGTYSGLLECGAD-IMKTGPLGFFKGLVPAFIRLGPQTILTFVFLEQLR 866 PG Y +C + + GP F+KG P+F+RLG ++ FV EQ++ Sbjct: 249 PGQYHSPFDCMLKMVTQEGPTAFYKGFTPSFLRLGSWNVVMFVTYEQMK 297
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 120,772,616
Number of Sequences: 369166
Number of extensions: 2485862
Number of successful extensions: 6744
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 5826
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6363
length of database: 68,354,980
effective HSP length: 112
effective length of database: 47,664,660
effective search space used: 11439518400
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail