DrC_00887
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00887 (1204 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q02874|H2AY_RAT Core histone macro-H2A.1 (Histone macroH... 223 7e-58 sp|O75367|H2AY_HUMAN Core histone macro-H2A.1 (Histone macr... 219 2e-56 sp|Q9P0M6|H2AW_HUMAN Core histone macro-H2A.2 (Histone macr... 202 2e-51 sp|Q8RB30|Y995_THETN Hypothetical UPF0189 protein TTE0995 101 5e-21 sp|P67341|YMDB_SALTY Hypothetical UPF0189 protein ymdB >gi|... 97 7e-20 sp|P27325|H2A_URECA Histone H2A 93 1e-18 sp|P69139|H2A3_PSAMI Late histone H2A.3, gonadal >gi|597995... 93 2e-18 sp|Q72M93|Y3295_LEPIC Hypothetical UPF0189 protein LIC13295 93 2e-18 sp|Q8EYT0|Y4133_LEPIN Hypothetical UPF0189 protein LA4133 93 2e-18 sp|P02269|H2A_ASTRU Histone H2A 92 2e-18
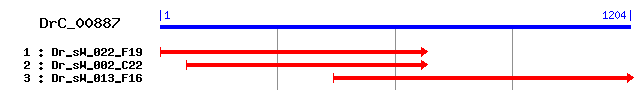
>sp|Q02874|H2AY_RAT Core histone macro-H2A.1 (Histone macroH2A1) (mH2A1) (H2A.y) (H2A/y) Length = 371 Score = 223 bits (569), Expect = 7e-58 Identities = 136/366 (37%), Positives = 198/366 (54%), Gaps = 12/366 (3%) Frame = +2 Query: 11 KKGFTKSKSQKAGLIFPVSRFHRYLKKQFPSSFRFKISIGAAVYAASVLEYLVAEIAEQA 190 KK S+S KAG+IFPV R RY+KK P +++I +GA VY A+VLEYL AEI E A Sbjct: 8 KKSTKTSRSAKAGVIFPVGRMLRYIKKGHP---KYRIGVGAPVYMAAVLEYLTAEILELA 64 Query: 191 GNAARILKVKRINPRHIMLAISQDMEMNQALKHVILPSTGRPLNLPGK--PSKQNTKTEL 364 GNAAR K R+ PRHI+LA++ D E+NQ LK V + S G N+ + K+ +K +L Sbjct: 65 GNAARDNKKGRVTPRHILLAVANDEELNQLLKGVTIASGGVLPNIHPELLAKKRGSKGKL 124 Query: 365 KASII-------KRXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXSQSNSLDV 523 +A I K + ++ V Sbjct: 125 EAIITPPPAKKAKSPSQKKPVAKKTGGKKGARKSKKQGEVSKAASADSTTEGAPTDGFTV 184 Query: 524 LNERTLFLGQKLVVVQGDIVEMSA---DAVVHPTGGSYGFGGEVGCALSNKGGAELTNEV 694 L+ ++LFLGQKL ++ +I ++ +A+++PT ++G L KGG E V Sbjct: 185 LSTKSLFLGQKLNLIHSEISNLAGFEVEAIINPTNADIDLKDDLGSTLEKKGGKEFVEAV 244 Query: 695 NKLRATKPSIKVCEAAVSGAVNIPYKYIIHVNSPSWTSNTNTAIANLEKAINNILDAADN 874 +LR ++V AAVS +P K++IH NSP W S+ + LEK + N L AD+ Sbjct: 245 LELRKKNGPLEVAGAAVSAGHGLPAKFVIHCNSPVWGSDKCEEL--LEKTVKNCLALADD 302 Query: 875 FNIQSLALPSISSGGGGYPKQIAAETILRTINSYFAGVAKSNIKQIYFVIYDKETLDVYK 1054 ++S+A PSI SG G+PKQ AA+ IL+ I+SYF S+IK +YFV++D E++ +Y Sbjct: 303 RKLKSIAFPSIGSGRNGFPKQTAAQLILKAISSYFVSTMSSSIKTVYFVLFDSESIGIYV 362 Query: 1055 AELTKL 1072 E+ KL Sbjct: 363 QEMAKL 368
>sp|O75367|H2AY_HUMAN Core histone macro-H2A.1 (Histone macroH2A1) (mH2A1) (H2A.y) (H2A/y) Length = 372 Score = 219 bits (557), Expect = 2e-56 Identities = 135/367 (36%), Positives = 198/367 (53%), Gaps = 13/367 (3%) Frame = +2 Query: 11 KKGFTKSKSQKAGLIFPVSRFHRYLKKQFPSSFRFKISIGAAVYAASVLEYLVAEIAEQA 190 KK S+S KAG+IFPV R RY+KK P +++I +GA VY A+VLEYL AEI E A Sbjct: 8 KKSTKTSRSAKAGVIFPVGRMLRYIKKGHP---KYRIGVGAPVYMAAVLEYLTAEILELA 64 Query: 191 GNAARILKVKRINPRHIMLAISQDMEMNQALKHVILPSTGRPLNLPGK--PSKQNTKTEL 364 GNAAR K R+ PRHI+LA++ D E+NQ LK V + S G N+ + K+ +K +L Sbjct: 65 GNAARDNKKGRVTPRHILLAVANDEELNQLLKGVTIASGGVLPNIHPELLAKKRGSKGKL 124 Query: 365 KASIIKRXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXSQSNSLD-------- 520 +A I S ++ + Sbjct: 125 EAIITPPPAKKAKSPSQKKPVSKKAGGKKGARKSKKKQGEVSKAASADSTTEGTPADGFT 184 Query: 521 VLNERTLFLGQKLVVVQGDIVEMSA---DAVVHPTGGSYGFGGEVGCALSNKGGAELTNE 691 VL+ ++LFLGQKL ++ +I ++ +A+++PT ++G L KGG E Sbjct: 185 VLSTKSLFLGQKLNLIHSEISNLAGFEVEAIINPTNADIDPKDDLGNTLEKKGGKEFVEA 244 Query: 692 VNKLRATKPSIKVCEAAVSGAVNIPYKYIIHVNSPSWTSNTNTAIANLEKAINNILDAAD 871 V +LR ++V AAVS +P K++IH NSP W ++ + LEK + N L AD Sbjct: 245 VLELRKKNGPLEVAGAAVSAGHGLPAKFVIHCNSPVWGADKCEEL--LEKTVKNCLALAD 302 Query: 872 NFNIQSLALPSISSGGGGYPKQIAAETILRTINSYFAGVAKSNIKQIYFVIYDKETLDVY 1051 + ++S+A PSI SG G+PKQ AA+ IL+ I+SYF S+IK +YFV++D E++ +Y Sbjct: 303 DKKLKSIAFPSIGSGRNGFPKQTAAQLILKAISSYFVSTMSSSIKTVYFVLFDSESIGIY 362 Query: 1052 KAELTKL 1072 E+ KL Sbjct: 363 VQEMAKL 369
>sp|Q9P0M6|H2AW_HUMAN Core histone macro-H2A.2 (Histone macroH2A2) (mH2A2) Length = 372 Score = 202 bits (514), Expect = 2e-51 Identities = 129/367 (35%), Positives = 191/367 (52%), Gaps = 13/367 (3%) Frame = +2 Query: 11 KKGFTKSKSQKAGLIFPVSRFHRYLKKQFPSSFRFKISIGAAVYAASVLEYLVAEIAEQA 190 KK S+S +AG+IFPV R RYLKK +F+++IS+GA VY A+V+EYL AEI E A Sbjct: 8 KKMSKLSRSARAGVIFPVGRLMRYLKK---GTFKYRISVGAPVYMAAVIEYLAAEILELA 64 Query: 191 GNAARILKVKRINPRHIMLAISQDMEMNQALKHVILPSTG-RPLNLPGKPSKQNTKTELK 367 GNAAR K RI PRHI+LA++ D E+NQ LK V + S G P P +K+ Sbjct: 65 GNAARDNKKARIAPRHILLAVANDEELNQLLKGVTIASGGVLPRIHPELLAKKRGTKGKS 124 Query: 368 ASIIKRXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXXSQSNSLD--------- 520 +I+ SNS Sbjct: 125 ETILSPPPEKRGRKATSGKKGGKKSKAAKPRTSKKSKPKDSDKEGTSNSTSEDGPGDGFT 184 Query: 521 VLNERTLFLGQKLVVVQGDIVE---MSADAVVHPTGGSYGFGGEVGCALSNKGGAELTNE 691 +L+ ++L LGQKL + Q DI M + +VHPT ++G AL GG E Sbjct: 185 ILSSKSLVLGQKLSLTQSDISHIGSMRVEGIVHPTTAEIDLKEDIGKALEKAGGKEFLET 244 Query: 692 VNKLRATKPSIKVCEAAVSGAVNIPYKYIIHVNSPSWTSNTNTAIANLEKAINNILDAAD 871 V +LR ++ ++V EAAVS + + K++IH + P W S+ LE+ I N L AA+ Sbjct: 245 VKELRKSQGPLEVAEAAVSQSSGLAAKFVIHCHIPQWGSDK--CEEQLEETIKNCLSAAE 302 Query: 872 NFNIQSLALPSISSGGGGYPKQIAAETILRTINSYFAGVAKSNIKQIYFVIYDKETLDVY 1051 + ++S+A P SG +PKQ AA+ L+ I+++F + S++K +YF+++D E++ +Y Sbjct: 303 DKKLKSVAFPPFPSGRNCFPKQTAAQVTLKAISAHFDDSSASSLKNVYFLLFDSESIGIY 362 Query: 1052 KAELTKL 1072 E+ KL Sbjct: 363 VQEMAKL 369
>sp|Q8RB30|Y995_THETN Hypothetical UPF0189 protein TTE0995 Length = 175 Score = 101 bits (251), Expect = 5e-21 Identities = 54/174 (31%), Positives = 90/174 (51%) Frame = +2 Query: 551 QKLVVVQGDIVEMSADAVVHPTGGSYGFGGEVGCALSNKGGAELTNEVNKLRATKPSIKV 730 +K+ +++G+IV+ DA+V+ S GG V A+ GG + E+ +R + Sbjct: 3 EKIKLIKGNIVDQEVDAIVNAANSSLIGGGGVDGAIHKAGGPAIAEELKVIREKQGGCPT 62 Query: 731 CEAAVSGAVNIPYKYIIHVNSPSWTSNTNTAIANLEKAINNILDAADNFNIQSLALPSIS 910 A ++GA N+ KY+IH P W + L A L AD +N++++A PSIS Sbjct: 63 GHAVITGAGNLKAKYVIHAVGPIWKGGNHNEDNLLASAYIESLKLADEYNVKTIAFPSIS 122 Query: 911 SGGGGYPKQIAAETILRTINSYFAGVAKSNIKQIYFVIYDKETLDVYKAELTKL 1072 +G G+P + AA LR ++ Y G S+IK++ FV++ +VY +L Sbjct: 123 TGAYGFPVERAARIALRVVSDYLEG---SSIKEVRFVLFSDRDYEVYSKAYEEL 173
>sp|P67341|YMDB_SALTY Hypothetical UPF0189 protein ymdB sp|P67342|YMDB_SALTI Hypothetical UPF0189 protein ymdB Length = 179 Score = 97.4 bits (241), Expect = 7e-20 Identities = 57/172 (33%), Positives = 90/172 (52%) Frame = +2 Query: 554 KLVVVQGDIVEMSADAVVHPTGGSYGFGGEVGCALSNKGGAELTNEVNKLRATKPSIKVC 733 +L V+QGDI ++S DA+V+ S GG V A+ G L + +R + + Sbjct: 4 RLQVIQGDITQLSVDAIVNAANASLMGGGGVDGAIHRAAGPALLDACKLIRQQQGECQTG 63 Query: 734 EAAVSGAVNIPYKYIIHVNSPSWTSNTNTAIANLEKAINNILDAADNFNIQSLALPSISS 913 A ++ A + K +IH P W + LE+A N L A+ + +S+A P+IS+ Sbjct: 64 HAVITPAGKLSAKAVIHTVGPVWRGGEHQEAELLEEAYRNCLLLAEANHFRSIAFPAIST 123 Query: 914 GGGGYPKQIAAETILRTINSYFAGVAKSNIKQIYFVIYDKETLDVYKAELTK 1069 G GYP+ AAE +RT++ + A +Q+YFV YD+ET +Y LT+ Sbjct: 124 GVYGYPRAQAAEVAVRTVSDFITRYALP--EQVYFVCYDEETARLYARLLTQ 173
>sp|P27325|H2A_URECA Histone H2A Length = 125 Score = 93.2 bits (230), Expect = 1e-18 Identities = 56/118 (47%), Positives = 71/118 (60%), Gaps = 4/118 (3%) Frame = +2 Query: 14 KGFTKSKSQKAGLIFPVSRFHRYLKKQFPSSFRFKISIGAAVYAASVLEYLVAEIAEQAG 193 KG +KS+S +AGL FPV R HR L+K ++ +I GA VY A+V+EYL AE+ E AG Sbjct: 11 KGKSKSRSSRAGLQFPVGRIHRLLRK---GNYAERIGAGAPVYLAAVMEYLAAEVLELAG 67 Query: 194 NAARILKVKRINPRHIMLAISQDMEMNQALKHVILPSTGRPLNLPG----KPSKQNTK 355 NAAR K RI PRH+ LAI D E+N+ L V + G N+ K S Q TK Sbjct: 68 NAARDNKKTRIIPRHLQLAIRNDEELNKLLSGVTIAQGGVLPNIQAVLLPKKSSQKTK 125
>sp|P69139|H2A3_PSAMI Late histone H2A.3, gonadal sp|P69140|H2A_PARAN Histone H2A, gonadal Length = 126 Score = 92.8 bits (229), Expect = 2e-18 Identities = 55/119 (46%), Positives = 73/119 (61%), Gaps = 5/119 (4%) Frame = +2 Query: 14 KGFTKSKSQKAGLIFPVSRFHRYLKKQFPSSFRFKISIGAAVYAASVLEYLVAEIAEQAG 193 KG KS+S +AGL FPV R HR+L+K ++ ++ GA VY A+VLEYL AEI E AG Sbjct: 11 KGKAKSRSSRAGLQFPVGRVHRFLRK---GNYANRVGAGAPVYLAAVLEYLAAEILELAG 67 Query: 194 NAARILKVKRINPRHIMLAISQDMEMNQALKHVILPSTG-----RPLNLPGKPSKQNTK 355 NAAR K RI PRH+ LAI D E+N+ L V + G + + LP K +++K Sbjct: 68 NAARDNKKTRIIPRHLQLAIRNDEELNKLLGGVTIAQGGVLPNIQAVLLPKKTGSKSSK 126
>sp|Q72M93|Y3295_LEPIC Hypothetical UPF0189 protein LIC13295 Length = 175 Score = 92.8 bits (229), Expect = 2e-18 Identities = 50/169 (29%), Positives = 90/169 (53%) Frame = +2 Query: 545 LGQKLVVVQGDIVEMSADAVVHPTGGSYGFGGEVGCALSNKGGAELTNEVNKLRATKPSI 724 + K+ +++ DI ++ DA+V+ S GG V A+ GG E+ E K+R + Sbjct: 1 MNNKIKLIKEDITQLEVDAIVNAANSSLLGGGGVDGAIHRAGGPEILEECYKIREKQGEC 60 Query: 725 KVCEAAVSGAVNIPYKYIIHVNSPSWTSNTNTAIANLEKAINNILDAADNFNIQSLALPS 904 KV EA ++ A + K+IIH P W+ L A N L A N +++++A P+ Sbjct: 61 KVGEAVITTAGRLNAKFIIHTVGPIWSGGNKNEDELLSNAYKNSLLLAKNHSLKTIAFPN 120 Query: 905 ISSGGGGYPKQIAAETILRTINSYFAGVAKSNIKQIYFVIYDKETLDVY 1051 IS+G +PK+ AA+ ++++ + + I+ ++FV +D E L++Y Sbjct: 121 ISTGIYHFPKERAAKIAIQSVTKFLK--QDNQIQTVFFVCFDFENLEIY 167
>sp|Q8EYT0|Y4133_LEPIN Hypothetical UPF0189 protein LA4133 Length = 175 Score = 92.8 bits (229), Expect = 2e-18 Identities = 50/169 (29%), Positives = 90/169 (53%) Frame = +2 Query: 545 LGQKLVVVQGDIVEMSADAVVHPTGGSYGFGGEVGCALSNKGGAELTNEVNKLRATKPSI 724 + K+ +++ DI ++ DA+V+ S GG V A+ GG E+ E K+R + Sbjct: 1 MNNKIKLIKEDITQLEVDAIVNAANSSLLGGGGVDGAIHRAGGPEILEECYKIREKQGEC 60 Query: 725 KVCEAAVSGAVNIPYKYIIHVNSPSWTSNTNTAIANLEKAINNILDAADNFNIQSLALPS 904 KV EA ++ A + K+IIH P W+ L A N L A N +++++A P+ Sbjct: 61 KVGEAVITTAGRLNAKFIIHTVGPIWSGGNKNEDELLSNAYKNSLLLAKNHSLKTIAFPN 120 Query: 905 ISSGGGGYPKQIAAETILRTINSYFAGVAKSNIKQIYFVIYDKETLDVY 1051 IS+G +PK+ AA+ ++++ + + I+ ++FV +D E L++Y Sbjct: 121 ISTGIYHFPKERAAKIAIQSVTEFLK--QDNQIQTVFFVCFDFENLEIY 167
>sp|P02269|H2A_ASTRU Histone H2A Length = 124 Score = 92.4 bits (228), Expect = 2e-18 Identities = 53/111 (47%), Positives = 70/111 (63%), Gaps = 5/111 (4%) Frame = +2 Query: 26 KSKSQKAGLIFPVSRFHRYLKKQFPSSFRFKISIGAAVYAASVLEYLVAEIAEQAGNAAR 205 KS+S +AGL FPV R HR+L+K ++ ++ GA VY A+V+EYL AEI E AGNAAR Sbjct: 14 KSRSSRAGLQFPVGRVHRFLRK---GNYAERVGAGAPVYLAAVMEYLAAEILELAGNAAR 70 Query: 206 ILKVKRINPRHIMLAISQDMEMNQALKHVILPSTG-----RPLNLPGKPSK 343 K RINPRH+ LAI D E+N+ L V + G + + LP K +K Sbjct: 71 DNKKTRINPRHLQLAIRNDEELNKLLSGVTIAQGGVLPNIQAVLLPKKTAK 121
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 115,862,075
Number of Sequences: 369166
Number of extensions: 2127537
Number of successful extensions: 6384
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 5949
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6245
length of database: 68,354,980
effective HSP length: 113
effective length of database: 47,479,925
effective search space used: 13626738475
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail