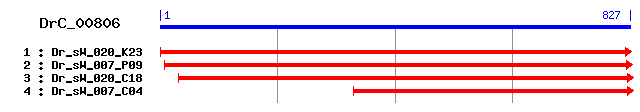
DrC_00806
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00806 (827 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|O70351|HCD2_RAT 3-hydroxyacyl-CoA dehydrogenase type II ... 258 1e-68 sp|O02691|HCD2_BOVIN 3-hydroxyacyl-CoA dehydrogenase type I... 254 2e-67 sp|O18404|HCD2_DROME 3-hydroxyacyl-CoA dehydrogenase type I... 253 6e-67 sp|Q99714|HCD2_HUMAN 3-hydroxyacyl-CoA dehydrogenase type I... 252 8e-67 sp|O08756|HCD2_MOUSE 3-hydroxyacyl-CoA dehydrogenase type I... 250 4e-66 sp|Q9X248|FABG_THEMA 3-oxoacyl-[acyl-carrier-protein] reduc... 95 3e-19 sp|P99120|BUTA_STAAN Acetoin(diacetyl) reductase (Acetoin d... 90 9e-18 sp|P46331|YXBG_BACSU Hypothetical oxidoreductase yxbG 86 2e-16 sp|P51831|FABG_BACSU 3-oxoacyl-[acyl-carrier-protein] reduc... 85 3e-16 sp|P66781|YD50_MYCTU Putative oxidoreductase Rv1350/MT1393 ... 84 5e-16
>sp|O70351|HCD2_RAT 3-hydroxyacyl-CoA dehydrogenase type II (Type II HADH) (Endoplasmic reticulum-associated amyloid beta-peptide binding protein) Length = 261 Score = 258 bits (660), Expect = 1e-68 Identities = 126/236 (53%), Positives = 166/236 (70%), Gaps = 6/236 (2%) Frame = +2 Query: 77 TVERLTEQGCKVAICDIQ------EPKKLPANSYYIKTDVSCANDVENAITQIKSEAKKL 238 T +RL QG + D+ E KKL N + +V+ +V+ A+T K + ++ Sbjct: 26 TAKRLVGQGATAVLLDVPNSEGETEAKKLGGNCIFAPANVTSEKEVQAALTLAKEKFGRI 85 Query: 239 DILVNCAGIGVAVKVYNRKKNSPHPLDTFERIVKVNAAGTFNMIRLCCSMFNSNEPNEFN 418 D+ VNCAGI VA+K Y+ KKN H L+ F+R++ VN GTFN+IRL + NEP++ Sbjct: 86 DVAVNCAGIAVAIKTYHEKKNQVHTLEDFQRVINVNLIGTFNVIRLVAGVMGQNEPDQGG 145 Query: 419 QRGIVINTSSIAAFEGQCGQSAYSASQGAIVAMTLPLARDLVAMGVRVMTIAPGIFLTPL 598 QRG++INT+S+AAFEGQ GQ+AYSAS+G IV MTLP+ARDL +G+RV+TIAPG+F TPL Sbjct: 146 QRGVIINTASVAAFEGQVGQAAYSASKGGIVGMTLPIARDLAPIGIRVVTIAPGLFATPL 205 Query: 599 LENVPDSARLNLSKIVPFPSRLGHPDEFAHLVQSIIENPMLNGEVIRIDGALRMMP 766 L +PD R L+ VPFPSRLG P E+AHLVQ +IENP LNGEVIR+DGA+RM P Sbjct: 206 LTTLPDKVRNFLASQVPFPSRLGDPAEYAHLVQMVIENPFLNGEVIRLDGAIRMQP 261
>sp|O02691|HCD2_BOVIN 3-hydroxyacyl-CoA dehydrogenase type II (Type II HADH) Length = 261 Score = 254 bits (649), Expect = 2e-67 Identities = 128/236 (54%), Positives = 164/236 (69%), Gaps = 6/236 (2%) Frame = +2 Query: 77 TVERLTEQGCKVAICDIQ------EPKKLPANSYYIKTDVSCANDVENAITQIKSEAKKL 238 T ERL QG + D+ + KKL + + DV+ DV+ A+T + + ++ Sbjct: 26 TAERLVGQGATAVLLDLPNSDGETQAKKLGKSCAFAPADVTSEKDVQAALTLAREKFGRV 85 Query: 239 DILVNCAGIGVAVKVYNRKKNSPHPLDTFERIVKVNAAGTFNMIRLCCSMFNSNEPNEFN 418 D+ VNCAGI VA K YN KK+ H L+ F+R++ VN GTFN+IRL NEP++ Sbjct: 86 DVAVNCAGIAVASKTYNLKKSQAHTLEDFQRVINVNLIGTFNVIRLVAGEMGQNEPDQGG 145 Query: 419 QRGIVINTSSIAAFEGQCGQSAYSASQGAIVAMTLPLARDLVAMGVRVMTIAPGIFLTPL 598 QRG++INT+S+AAFEGQ GQ+AYSAS+G IV MTLP+ARDL MG+RVMTIAPG+F TPL Sbjct: 146 QRGVIINTASVAAFEGQVGQAAYSASKGGIVGMTLPIARDLAPMGIRVMTIAPGLFGTPL 205 Query: 599 LENVPDSARLNLSKIVPFPSRLGHPDEFAHLVQSIIENPMLNGEVIRIDGALRMMP 766 L +PD R L+ VPFPSRLG P E+AHLVQ+IIEN LNGEVIR+DGA+RM P Sbjct: 206 LTTLPDKVRNFLASQVPFPSRLGDPAEYAHLVQAIIENSFLNGEVIRLDGAIRMQP 261
>sp|O18404|HCD2_DROME 3-hydroxyacyl-CoA dehydrogenase type II (Type II HADH) (Scully protein) Length = 255 Score = 253 bits (645), Expect = 6e-67 Identities = 121/236 (51%), Positives = 164/236 (69%), Gaps = 6/236 (2%) Frame = +2 Query: 77 TVERLTEQGCKVAICDI------QEPKKLPANSYYIKTDVSCANDVENAITQIKSEAKKL 238 T ERL +QG V + D+ + K+L ++ DV+ DV A+ K + +L Sbjct: 20 TAERLAKQGASVILADLPSSKGNEVAKELGDKVVFVPVDVTSEKDVSAALQTAKDKFGRL 79 Query: 239 DILVNCAGIGVAVKVYNRKKNSPHPLDTFERIVKVNAAGTFNMIRLCCSMFNSNEPNEFN 418 D+ VNCAG AVK +N KN H L+ F+R++ +N GTFN+IRL + +NEPN+ Sbjct: 80 DLTVNCAGTATAVKTFNFNKNVAHRLEDFQRVININTVGTFNVIRLSAGLMGANEPNQDG 139 Query: 419 QRGIVINTSSIAAFEGQCGQSAYSASQGAIVAMTLPLARDLVAMGVRVMTIAPGIFLTPL 598 QRG+++NT+S+AAF+GQ GQ+AYSAS+ A+V MTLP+ARDL G+R+ TIAPG+F TP+ Sbjct: 140 QRGVIVNTASVAAFDGQIGQAAYSASKAAVVGMTLPIARDLSTQGIRICTIAPGLFNTPM 199 Query: 599 LENVPDSARLNLSKIVPFPSRLGHPDEFAHLVQSIIENPMLNGEVIRIDGALRMMP 766 L +P+ R L+K +PFP RLG P E+AHLVQ+I ENP+LNGEVIRIDGALRMMP Sbjct: 200 LAALPEKVRTFLAKSIPFPQRLGEPSEYAHLVQAIYENPLLNGEVIRIDGALRMMP 255
>sp|Q99714|HCD2_HUMAN 3-hydroxyacyl-CoA dehydrogenase type II (Type II HADH) (Endoplasmic reticulum-associated amyloid beta-peptide binding protein) (Short-chain type dehydrogenase/reductase XH98G2) Length = 261 Score = 252 bits (644), Expect = 8e-67 Identities = 127/236 (53%), Positives = 163/236 (69%), Gaps = 6/236 (2%) Frame = +2 Query: 77 TVERLTEQGCKVAICDIQ------EPKKLPANSYYIKTDVSCANDVENAITQIKSEAKKL 238 T ERL QG + D+ + KKL N + DV+ DV+ A+ K + ++ Sbjct: 26 TAERLVGQGASAVLLDLPNSGGEAQAKKLGNNCVFAPADVTSEKDVQTALALAKGKFGRV 85 Query: 239 DILVNCAGIGVAVKVYNRKKNSPHPLDTFERIVKVNAAGTFNMIRLCCSMFNSNEPNEFN 418 D+ VNCAGI VA K YN KK H L+ F+R++ VN GTFN+IRL NEP++ Sbjct: 86 DVAVNCAGIAVASKTYNLKKGQTHTLEDFQRVLDVNLMGTFNVIRLVAGEMGQNEPDQGG 145 Query: 419 QRGIVINTSSIAAFEGQCGQSAYSASQGAIVAMTLPLARDLVAMGVRVMTIAPGIFLTPL 598 QRG++INT+S+AAFEGQ GQ+AYSAS+G IV MTLP+ARDL +G+RVMTIAPG+F TPL Sbjct: 146 QRGVIINTASVAAFEGQVGQAAYSASKGGIVGMTLPIARDLAPIGIRVMTIAPGLFGTPL 205 Query: 599 LENVPDSARLNLSKIVPFPSRLGHPDEFAHLVQSIIENPMLNGEVIRIDGALRMMP 766 L ++P+ L+ VPFPSRLG P E+AHLVQ+IIENP LNGEVIR+DGA+RM P Sbjct: 206 LTSLPEKVCNFLASQVPFPSRLGDPAEYAHLVQAIIENPFLNGEVIRLDGAIRMQP 261
>sp|O08756|HCD2_MOUSE 3-hydroxyacyl-CoA dehydrogenase type II (Type II HADH) (Endoplasmic reticulum-associated amyloid beta-peptide binding protein) Length = 261 Score = 250 bits (638), Expect = 4e-66 Identities = 122/236 (51%), Positives = 165/236 (69%), Gaps = 6/236 (2%) Frame = +2 Query: 77 TVERLTEQGCKVAICDI------QEPKKLPANSYYIKTDVSCANDVENAITQIKSEAKKL 238 T +RL QG + D+ + KKL + + +V+ +++ A+T K + ++ Sbjct: 26 TAKRLVGQGATAVLLDVPDSEGESQAKKLGESCIFAPANVTSEKEIQAALTLAKEKFGRI 85 Query: 239 DILVNCAGIGVAVKVYNRKKNSPHPLDTFERIVKVNAAGTFNMIRLCCSMFNSNEPNEFN 418 D+ VNCAGI VA+K Y++KKN H L+ F+R++ VN GTFN+IRL NEP++ Sbjct: 86 DVAVNCAGIAVAIKTYHQKKNKIHTLEDFQRVINVNLIGTFNVIRLVAGEMGQNEPDQGG 145 Query: 419 QRGIVINTSSIAAFEGQCGQSAYSASQGAIVAMTLPLARDLVAMGVRVMTIAPGIFLTPL 598 QRG++INT+S+AAFEGQ GQ+AYSAS+G I MTLP+ARDL G+RV+TIAPG+F TPL Sbjct: 146 QRGVIINTASVAAFEGQVGQAAYSASKGGIDGMTLPIARDLAPTGIRVVTIAPGLFATPL 205 Query: 599 LENVPDSARLNLSKIVPFPSRLGHPDEFAHLVQSIIENPMLNGEVIRIDGALRMMP 766 L +P+ R L+ VPFPSRLG P E+AHLVQ+IIENP LNGEVIR+DGA+RM P Sbjct: 206 LTTLPEKVRNFLASQVPFPSRLGDPAEYAHLVQTIIENPFLNGEVIRLDGAIRMQP 261
>sp|Q9X248|FABG_THEMA 3-oxoacyl-[acyl-carrier-protein] reductase (3-ketoacyl-acyl carrier protein reductase) Length = 246 Score = 94.7 bits (234), Expect = 3e-19 Identities = 59/213 (27%), Positives = 107/213 (50%), Gaps = 3/213 (1%) Frame = +2 Query: 125 IQEPKKLPANSYYIKTDVSCANDVENAITQIKSEAKKLDILVNCAGIGVAVKVYNRKKNS 304 ++E + LP +V+ + ++ + ++ + ++D+LVN AGI + K+ Sbjct: 46 VKEAEGLPGKVDPYVLNVTDRDQIKEVVEKVVQKYGRIDVLVNNAGITRDALLVRMKE-- 103 Query: 305 PHPLDTFERIVKVNAAGTFNMIRLCCSMFNSNEPNEFNQR-GIVINTSSIAAFEGQCGQS 481 + ++ ++ VN G FN+ ++ P QR G ++N SS+ G GQ+ Sbjct: 104 ----EDWDAVINVNLKGVFNVTQMVV-------PYMIKQRNGSIVNVSSVVGIYGNPGQT 152 Query: 482 AYSASQGAIVAMTLPLARDLVAMGVRVMTIAPGIFLTPLLENVPDSARLNLSKIVPFPSR 661 Y+AS+ ++ MT A++L +RV +APG TP+ E +P+ AR +P R Sbjct: 153 NYAASKAGVIGMTKTWAKELAGRNIRVNAVAPGFIETPMTEKLPEKARETALSRIPL-GR 211 Query: 662 LGHPDEFAHLVQSII--ENPMLNGEVIRIDGAL 754 G P+E A ++ + E+ + G+VI IDG L Sbjct: 212 FGKPEEVAQVILFLASDESSYVTGQVIGIDGGL 244
>sp|P99120|BUTA_STAAN Acetoin(diacetyl) reductase (Acetoin dehydrogenase) (AR) (Meso-2,3-butanediol dehydrogenase) sp|P66776|BUTA_STAAW Acetoin(diacetyl) reductase (Acetoin dehydrogenase) (AR) (Meso-2,3-butanediol dehydrogenase) sp|P66775|BUTA_STAAM Acetoin(diacetyl) reductase (Acetoin dehydrogenase) (AR) (Meso-2,3-butanediol dehydrogenase) sp|Q6GKH9|BUTA_STAAR Acetoin(diacetyl) reductase (Acetoin dehydrogenase) (AR) (Meso-2,3-butanediol dehydrogenase) sp|Q6GCZ8|BUTA_STAAS Acetoin(diacetyl) reductase (Acetoin dehydrogenase) (AR) (Meso-2,3-butanediol dehydrogenase) Length = 258 Score = 89.7 bits (221), Expect = 9e-18 Identities = 71/251 (28%), Positives = 114/251 (45%), Gaps = 26/251 (10%) Frame = +2 Query: 83 ERLTEQGCKVAICDIQEPKKLPA---------NSYYIKTDVSCANDVENAITQIKSEAKK 235 ERL E G KVA+ D E A + IK DVS +DV NA+ Q ++ Sbjct: 22 ERLVEDGFKVAVVDFNEEGAKAAALKLSSDGTKAIAIKADVSNRDDVFNAVRQTAAQFGD 81 Query: 236 LDILVNCAGIGVAVKVYNRKKNSPHPLDT-----FERIVKVNAAGTFNMIRLCCSMFNSN 400 ++VN AG+G P+DT F+ + VN AG I+ F Sbjct: 82 FHVMVNNAGLGPTT-----------PIDTITEEQFKTVYGVNVAGVLWGIQAAHEQFK-- 128 Query: 401 EPNEFNQRGIVINTSSIAAFEGQCGQSAYSASQGAIVAMTLPLARDLVAMGVRVMTIAPG 580 +FN G +IN +S A EG G S Y +++ A+ +T A+DL + G+ V APG Sbjct: 129 ---KFNHGGKIINATSQAGVEGNPGLSLYCSTKFAVRGLTQVAAQDLASEGITVNAFAPG 185 Query: 581 IFLTPLLENVPDSARLNLSKIVPFP----------SRLGHPDEFAHLVQSII--ENPMLN 724 I TP++E++ + K + R+ P++ +++V + ++ + Sbjct: 186 IVQTPMMESIAVATAEEAGKPEAWGWEQFTSQIALGRVSQPEDVSNVVSFLAGKDSDYIT 245 Query: 725 GEVIRIDGALR 757 G+ I +DG +R Sbjct: 246 GQTIIVDGGMR 256
>sp|P46331|YXBG_BACSU Hypothetical oxidoreductase yxbG Length = 273 Score = 85.5 bits (210), Expect = 2e-16 Identities = 76/265 (28%), Positives = 114/265 (43%), Gaps = 19/265 (7%) Frame = +2 Query: 77 TVERLTEQGCKVAICDIQEP---------KKLPANSYYIKTDVSCANDVENAITQIKSEA 229 T E +G +V I DI + +K + DVS N V+ QIK Sbjct: 22 TAEVFANEGARVIIGDINKDQMEETVDAIRKNGGQAESFHLDVSDENSVKAFADQIKDAC 81 Query: 230 KKLDILVNCAGIGVAVKVYNRKKNSPHPLDTFERIVKVNAAGTFNMIRLCCSMFNSNEPN 409 +DIL N AG+ K +P+D F+RI+ V+ GTF CS + P Sbjct: 82 GTIDILFNNAGVD-----QEGGKVHEYPVDLFDRIIAVDLRGTF-----LCSKYLI--PL 129 Query: 410 EFNQRGIVINTSSIAAFEGQCGQSAYSASQGAIVAMTLPLARDLVAMGVRVMTIAPGIFL 589 G +INTSS++ +S Y+A++G I +T +A D G+RV +I+PG Sbjct: 130 MLENGGSIINTSSMSGRAADLDRSGYNAAKGGITNLTKAMAIDYARNGIRVNSISPGTIE 189 Query: 590 TPLLENVPDSARLNLSK--------IVPFPSRLGHPDEFAHLVQSII--ENPMLNGEVIR 739 TPL++ + + + + I P RLG P E A + + ++ + GE I Sbjct: 190 TPLIDKLAGTKEQEMGEQFREANKWITPL-GRLGQPKEMATVALFLASDDSSYVTGEDIT 248 Query: 740 IDGALRMMP*LLFWYNKTCIRFKMK 814 DG + W K I K K Sbjct: 249 ADGGIMAYT----WPGKMLIEEKWK 269
>sp|P51831|FABG_BACSU 3-oxoacyl-[acyl-carrier-protein] reductase (3-ketoacyl-acyl carrier protein reductase) Length = 246 Score = 84.7 bits (208), Expect = 3e-16 Identities = 56/214 (26%), Positives = 98/214 (45%), Gaps = 2/214 (0%) Frame = +2 Query: 125 IQEPKKLPANSYYIKTDVSCANDVENAITQIKSEAKKLDILVNCAGIGVAVKVYNRKKNS 304 + E K + + +K DVS DV+N I + S +DILVN AGI + K+ Sbjct: 46 VDEIKSMGRKAIAVKADVSNPEDVQNMIKETLSVFSTIDILVNNAGITRDNLIMRMKE-- 103 Query: 305 PHPLDTFERIVKVNAAGTFNMIRLCCSMFNSNEPNEFNQRGIVINTSSIAAFEGQCGQSA 484 D ++ ++ +N G FN + + G +IN SSI G GQ+ Sbjct: 104 ----DEWDDVININLKGVFNCTKAVTRQMMKQ------RSGRIINVSSIVGVSGNPGQAN 153 Query: 485 YSASQGAIVAMTLPLARDLVAMGVRVMTIAPGIFLTPLLENVPDSARLNLSKIVPFPSRL 664 Y A++ ++ +T A++L + + V IAPG T + + + + + K +P +R Sbjct: 154 YVAAKAGVIGLTKSSAKELASRNITVNAIAPGFISTDMTDKLAKDVQDEMLKQIPL-ARF 212 Query: 665 GHPDEFAHLVQSIIEN--PMLNGEVIRIDGALRM 760 G P + + +V + + G+ + IDG + M Sbjct: 213 GEPSDVSSVVTFLASEGARYMTGQTLHIDGGMVM 246
>sp|P66781|YD50_MYCTU Putative oxidoreductase Rv1350/MT1393 sp|P66782|YD85_MYCBO Putative oxidoreductase Mb1385 Length = 247 Score = 84.0 bits (206), Expect = 5e-16 Identities = 63/233 (27%), Positives = 113/233 (48%), Gaps = 11/233 (4%) Frame = +2 Query: 83 ERLTEQGCKVAICDIQ------EPKKLPAN--SYYIKTDVSCANDVENAITQIKSEAKKL 238 +R +G +V + D+ K+L + + ++ DV+ A+DV+ I L Sbjct: 25 QRFVAEGARVVLGDVNLEATEVAAKRLGGDDVALAVRCDVTQADDVDILIRTAVERFGGL 84 Query: 239 DILVNCAGIGVAVKVYNRKKNSPHPLDTFERIVKVNAAGTFNMIRLCCSMFNSNEPNEFN 418 D++VN AGI + + + F++++ V+ GT+N RL ++ + Sbjct: 85 DVMVNNAGITRDATMRTMTE------EQFDQVIAVHLKGTWNGTRLAAAIMRERK----- 133 Query: 419 QRGIVINTSSIAAFEGQCGQSAYSASQGAIVAMTLPLARDLVAMGVRVMTIAPGIFLTPL 598 RG ++N SS++ G GQ+ YSA++ IV MT A++L +G+RV IAPG+ + + Sbjct: 134 -RGAIVNMSSVSGKVGMVGQTNYSAAKAGIVGMTKAAAKELAHLGIRVNAIAPGLIRSAM 192 Query: 599 LENVPDSARLNLSKIVPFP-SRLGHPDEFAHLVQSIIE--NPMLNGEVIRIDG 748 E +P R+ K+ P R G P E A + + + + G V+ + G Sbjct: 193 TEAMPQ--RIWDQKLAEVPMGRAGEPSEVASVAVFLASDLSSYMTGTVLDVTG 243
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 87,981,557
Number of Sequences: 369166
Number of extensions: 1793805
Number of successful extensions: 5383
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 5094
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5247
length of database: 68,354,980
effective HSP length: 109
effective length of database: 48,218,865
effective search space used: 8004331590
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail