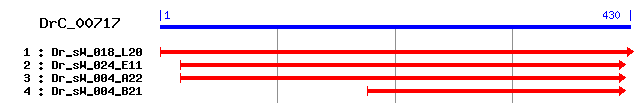
DrC_00717
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00717 (430 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q9XSK2|CD63_BOVIN CD63 antigen 43 2e-04 sp|O70401|TSN6_MOUSE Tetraspanin-6 (Tspan-6) (Transmembrane... 42 5e-04 sp|O43657|TSN6_HUMAN Tetraspanin-6 (Tspan-6) (Transmembrane... 40 0.003 sp|P08962|CD63_HUMAN CD63 antigen (Melanoma-associated anti... 38 0.008 sp|Q9QY33|TSN3_MOUSE Tetraspanin-3 (Tspan-3) (Transmembrane... 38 0.010 sp|O60637|TSN3_HUMAN Tetraspanin-3 (Tspan-3) (Transmembrane... 37 0.013 sp|P41731|CD63_MOUSE CD63 antigen 37 0.023 sp|P28648|CD63_RAT CD63 antigen (AD1 antigen) 36 0.039 sp|Q61451|CD53_MOUSE Leukocyte surface antigen CD53 (Cell s... 36 0.039 sp|Q62283|TSN7_MOUSE Tetraspanin-7 (Tspan-7) (Transmembrane... 35 0.050
>sp|Q9XSK2|CD63_BOVIN CD63 antigen Length = 237 Score = 43.1 bits (100), Expect = 2e-04 Identities = 20/77 (25%), Positives = 40/77 (51%), Gaps = 9/77 (11%) Frame = +3 Query: 15 IDELQNHFACCGSNSFSDWNSTVFYSNSTNYPSSC-----YSLGGSI----LHTAGCRSK 167 +D++Q F CCG+ +++DW + +N P SC ++ G + +HT GC K Sbjct: 136 LDKMQKDFECCGAANYTDWEKILAVTNKV--PDSCCVNITHNCGINFVVKDIHTEGCVEK 193 Query: 168 LYDWYRRFFIAILSVPI 218 + W R+ + +++ + Sbjct: 194 IAAWLRKNVLVVVAAAL 210
>sp|O70401|TSN6_MOUSE Tetraspanin-6 (Tspan-6) (Transmembrane 4 superfamily member 6) Length = 245 Score = 42.0 bits (97), Expect = 5e-04 Identities = 17/43 (39%), Positives = 25/43 (58%) Frame = +3 Query: 6 SRDIDELQNHFACCGSNSFSDWNSTVFYSNSTNYPSSCYSLGG 134 S +D++Q+ CCG ++ DW T +YS T +P SC L G Sbjct: 141 SEAVDKIQSTLHCCGVTNYGDWKGTNYYS-ETGFPKSCCKLEG 182
>sp|O43657|TSN6_HUMAN Tetraspanin-6 (Tspan-6) (Transmembrane 4 superfamily member 6) (T245 protein) (Tetraspanin TM4-D) (A15 homolog) Length = 245 Score = 39.7 bits (91), Expect = 0.003 Identities = 16/41 (39%), Positives = 22/41 (53%) Frame = +3 Query: 6 SRDIDELQNHFACCGSNSFSDWNSTVFYSNSTNYPSSCYSL 128 S +D++QN CCG + DW T +YS +P SC L Sbjct: 141 SHAVDKIQNTLHCCGVTDYRDWTDTNYYSEK-GFPKSCCKL 180
>sp|P08962|CD63_HUMAN CD63 antigen (Melanoma-associated antigen ME491) (Lysosome-associated membrane glycoprotein 3) (LAMP-3) (Ocular melanoma-associated antigen) (OMA81H) (Granulophysin) (Tetraspanin-30) (Tspan-30) Length = 238 Score = 38.1 bits (87), Expect = 0.008 Identities = 18/77 (23%), Positives = 33/77 (42%), Gaps = 9/77 (11%) Frame = +3 Query: 15 IDELQNHFACCGSNSFSDWNSTVFYSNSTNYPSSC---------YSLGGSILHTAGCRSK 167 +D +Q F CCG+ +++DW S + P SC + +H GC K Sbjct: 136 LDRMQADFKCCGAANYTDWEKIPSMSKN-RVPDSCCINVTVGCGINFNEKAIHKEGCVEK 194 Query: 168 LYDWYRRFFIAILSVPI 218 + W R+ + + + + Sbjct: 195 IGGWLRKNVLVVAAAAL 211
>sp|Q9QY33|TSN3_MOUSE Tetraspanin-3 (Tspan-3) (Transmembrane 4 superfamily member 8) (Tetraspanin TM4-A) (OSP-associated protein 1) (OAP-1) Length = 253 Score = 37.7 bits (86), Expect = 0.010 Identities = 22/65 (33%), Positives = 33/65 (50%), Gaps = 12/65 (18%) Frame = +3 Query: 6 SRDIDELQNHFACCGSNSFSDWNSTVFYSNSTN--YPSSC-----YSLGGSI-----LHT 149 SR ID +Q CCG +++SDW +T ++ + N P SC S GS+ L+ Sbjct: 135 SRAIDYVQRQLHCCGIHNYSDWENTDWFKETKNQSVPLSCCRETAKSCNGSLANPSDLYA 194 Query: 150 AGCRS 164 GC + Sbjct: 195 EGCEA 199
>sp|O60637|TSN3_HUMAN Tetraspanin-3 (Tspan-3) (Transmembrane 4 superfamily member 8) (Tetraspanin TM4-A) Length = 253 Score = 37.4 bits (85), Expect = 0.013 Identities = 19/54 (35%), Positives = 29/54 (53%), Gaps = 7/54 (12%) Frame = +3 Query: 6 SRDIDELQNHFACCGSNSFSDWNSTVFYSNSTN--YPSSC-----YSLGGSILH 146 SR ID +Q CCG +++SDW +T ++ + N P SC + GS+ H Sbjct: 135 SRAIDYVQRQLHCCGIHNYSDWENTDWFKETKNQSVPLSCCRETASNCNGSLAH 188
>sp|P41731|CD63_MOUSE CD63 antigen Length = 238 Score = 36.6 bits (83), Expect = 0.023 Identities = 17/76 (22%), Positives = 35/76 (46%), Gaps = 8/76 (10%) Frame = +3 Query: 15 IDELQNHFACCGSNSFSDWNSTVFYSNSTNYPSSCYSL--------GGSILHTAGCRSKL 170 +D+LQ CCG+++++DW + + S C ++ S +HT GC + Sbjct: 136 LDKLQKENNCCGASNYTDWENIPGMAKDRVPDSCCINITVGCGNDFKESTIHTQGCVETI 195 Query: 171 YDWYRRFFIAILSVPI 218 W R+ + + + + Sbjct: 196 AIWLRKNILLVAAAAL 211
>sp|P28648|CD63_RAT CD63 antigen (AD1 antigen) Length = 238 Score = 35.8 bits (81), Expect = 0.039 Identities = 17/66 (25%), Positives = 30/66 (45%), Gaps = 8/66 (12%) Frame = +3 Query: 15 IDELQNHFACCGSNSFSDWNSTVFYSNSTNYPSSCYSL--------GGSILHTAGCRSKL 170 +D+LQ CCG+++++DW + S C ++ S +HT GC + Sbjct: 136 LDKLQKENKCCGASNYTDWERIPGMAKDRVPDSCCINITVGCGNDFKESTIHTQGCVETI 195 Query: 171 YDWYRR 188 W R+ Sbjct: 196 AAWLRK 201
>sp|Q61451|CD53_MOUSE Leukocyte surface antigen CD53 (Cell surface glycoprotein CD53) Length = 219 Score = 35.8 bits (81), Expect = 0.039 Identities = 22/62 (35%), Positives = 27/62 (43%) Frame = +3 Query: 18 DELQNHFACCGSNSFSDWNSTVFYSNSTNYPSSCYSLGGSILHTAGCRSKLYDWYRRFFI 197 D +Q CCG N SDW S PSSC S G + GC +K W+ F+ Sbjct: 136 DFIQTQLQCCGVNGSSDWTS--------GPPSSCPS-GADV---QGCYNKAKSWFHSNFL 183 Query: 198 AI 203 I Sbjct: 184 YI 185
>sp|Q62283|TSN7_MOUSE Tetraspanin-7 (Tspan-7) (Transmembrane 4 superfamily member 2) (Cell surface glycoprotein A15) (PE31) (TALLA homolog) Length = 249 Score = 35.4 bits (80), Expect = 0.050 Identities = 12/38 (31%), Positives = 24/38 (63%) Frame = +3 Query: 6 SRDIDELQNHFACCGSNSFSDWNSTVFYSNSTNYPSSC 119 SR +D +Q +CCG ++++W+S+ ++ + PS C Sbjct: 138 SRAVDHVQRSLSCCGVQNYTNWSSSPYFLDHGIPPSCC 175
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 48,101,119
Number of Sequences: 369166
Number of extensions: 921499
Number of successful extensions: 2429
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 2383
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 2425
length of database: 68,354,980
effective HSP length: 100
effective length of database: 49,881,480
effective search space used: 2095022160
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail