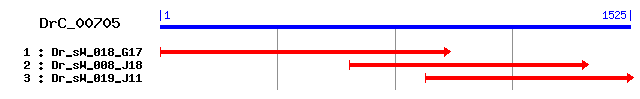
DrC_00705
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00705 (1525 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P12955|PEPD_HUMAN Xaa-Pro dipeptidase (X-Pro dipeptidase... 550 e-156 sp|Q11136|PEPD_MOUSE Xaa-Pro dipeptidase (X-Pro dipeptidase... 548 e-155 sp|P43590|YFH6_YEAST Hypothetical 61.8 kDa peptidase in MPR... 213 1e-54 sp|P15034|AMPP_ECOLI Xaa-Pro aminopeptidase (X-Pro aminopep... 182 2e-45 sp|P44881|AMPP_HAEIN Xaa-Pro aminopeptidase (X-Pro aminopep... 179 2e-44 sp|P0A3Z2|AMPP1_STRLI Xaa-Pro aminopeptidase I (X-Pro amino... 129 2e-29 sp|P0A3Z4|AMPP2_STRLI Xaa-Pro aminopeptidase II (X-Pro amin... 120 1e-26 sp|P21165|PEPQ_ECOLI Xaa-Pro dipeptidase (X-Pro dipeptidase... 118 5e-26 sp|P81535|PEPQ_PYRFU Xaa-Pro dipeptidase (X-Pro dipeptidase... 108 5e-23 sp|P40051|YEQ8_YEAST Hypothetical 58.0 kDa peptidase in PTP... 101 7e-21
>sp|P12955|PEPD_HUMAN Xaa-Pro dipeptidase (X-Pro dipeptidase) (Proline dipeptidase) (Prolidase) (Imidodipeptidase) Length = 493 Score = 550 bits (1417), Expect = e-156 Identities = 274/478 (57%), Positives = 347/478 (72%), Gaps = 1/478 (0%) Frame = +1 Query: 16 PAYQLG-DALSVPMTLHTENRKRLCAKMKATNKINNNAFILLQGGTDHYLGGSDSALTFR 192 P++ LG + L VP+ L NR+RLC +++ + + ++LQGG + +D+ + FR Sbjct: 7 PSFWLGNETLKVPLALFALNRQRLCERLRKNPAVQAGSIVVLQGGEETQRYCTDTGVLFR 66 Query: 193 QESYFHWAFGVLEPDFYGLIHIDSTKTILFMPRLDERYAIFMGALETKDDVKQKYAVDEV 372 QES+FHWAFGV EP YG+I +D+ K+ LF+PRL +A +MG + +K+ K+KYAVD+V Sbjct: 67 QESFFHWAFGVTEPGCYGVIDVDTGKSTLFVPRLPASHATWMGKIHSKEHFKEKYAVDDV 126 Query: 373 YYTDEILNVLEELKPSEIHTLHGLNTDSNNYTMEASFTGIANFQVINSVLHNEITECRVI 552 Y DEI +VL KPS + TL G+NTDS + EASF GI+ F+V N++LH EI ECRV Sbjct: 127 QYVDEIASVLTSQKPSVLLTLRGVNTDSGSVCREASFDGISKFEVNNTILHPEIVECRVF 186 Query: 553 KTNQELEVLRYANKASSKAHRDIMRSVHPGMYEYQIESLFLHHCYFHGAMRNVAYTSIGA 732 KT+ ELEVLRY NK SS+AHR++M++V GM EY++ESLF H+CY G MR+ +YT I Sbjct: 187 KTDMELEVLRYTNKISSEAHREVMKAVKVGMKEYELESLFEHYCYSRGGMRHSSYTCICG 246 Query: 733 TGCNCSVLHYGHAGAPNDRKIEDGDMCLFDMGGEYYCYSSDITCSFPVNGHFTKDQKIVY 912 +G N +VLHYGHAGAPNDR I++GDMCLFDMGGEYYC++SDITCSFP NG FT DQK VY Sbjct: 247 SGENSAVLHYGHAGAPNDRTIQNGDMCLFDMGGEYYCFASDITCSFPANGKFTADQKAVY 306 Query: 913 EAVLAASRAVKNACKPGVRWTDMHLLAERIILSSLKEAGLLQGDVNEMMAVRLGAVFMPH 1092 EAVL +SRAV A KPGV W DMH LA+RI L L G+L G V+ M+ LGAVFMPH Sbjct: 307 EAVLRSSRAVMGAMKPGVWWPDMHRLADRIHLEELAHMGILSGSVDAMVQAHLGAVFMPH 366 Query: 1093 GLGHLLGVDVHDVGGYIEDAIPRSELSGLRSLRTTQLLQENMELTVEPGCYFIDILLDAA 1272 GLGH LG+DVHDVGGY E + R + GLRSLRT + LQ M LTVEPG YFID LLD A Sbjct: 367 GLGHFLGIDVHDVGGYPE-GVERIDEPGLRSLRTARHLQPGMVLTVEPGIYFIDHLLDEA 425 Query: 1273 INNPEQARFMVINEINRFTRFGGVRIEDDIIVTSTGIECMTEVPRTCEEIEQRMAGND 1446 + +P +A F+ + RF FGGVRIE+D++VT +GIE +T VPRT EEIE MAG D Sbjct: 426 LADPARASFLNREVLQRFRGFGGVRIEEDVVVTDSGIELLTCVPRTVEEIEACMAGCD 483
>sp|Q11136|PEPD_MOUSE Xaa-Pro dipeptidase (X-Pro dipeptidase) (Proline dipeptidase) (Prolidase) (Imidodipeptidase) (Peptidase 4) Length = 493 Score = 548 bits (1411), Expect = e-155 Identities = 273/486 (56%), Positives = 350/486 (72%), Gaps = 1/486 (0%) Frame = +1 Query: 1 ARRVVPAYQLG-DALSVPMTLHTENRKRLCAKMKATNKINNNAFILLQGGTDHYLGGSDS 177 A V P++ LG + L VP+ L NR+RLC +++ + + ++LQGG + +D+ Sbjct: 2 ASTVRPSFSLGNETLKVPLALFALNRQRLCERLRKNGAVQAASAVVLQGGEEMQRYCTDT 61 Query: 178 ALTFRQESYFHWAFGVLEPDFYGLIHIDSTKTILFMPRLDERYAIFMGALETKDDVKQKY 357 ++ FRQES+FHWAFGV+E YG+I +D+ K+ LF+PRL + YA +MG + +K+ K+KY Sbjct: 62 SIIFRQESFFHWAFGVVESGCYGVIDVDTGKSTLFVPRLPDSYATWMGKIHSKEYFKEKY 121 Query: 358 AVDEVYYTDEILNVLEELKPSEIHTLHGLNTDSNNYTMEASFTGIANFQVINSVLHNEIT 537 AVD+V YTDEI +VL PS + TL G+NTDS + EASF GI+ F V N++LH EI Sbjct: 122 AVDDVQYTDEIASVLTSRNPSVLLTLRGVNTDSGSVCREASFEGISKFNVNNTILHPEIV 181 Query: 538 ECRVIKTNQELEVLRYANKASSKAHRDIMRSVHPGMYEYQIESLFLHHCYFHGAMRNVAY 717 ECRV KT+ ELEVLRY N+ SS+AHR++M++V GM EY++ESLF H+CY G MR+ +Y Sbjct: 182 ECRVFKTDMELEVLRYTNRISSEAHREVMKAVKVGMKEYEMESLFQHYCYSRGGMRHTSY 241 Query: 718 TSIGATGCNCSVLHYGHAGAPNDRKIEDGDMCLFDMGGEYYCYSSDITCSFPVNGHFTKD 897 T I +G N +VLHYGHAGAPNDR I+DGD+CLFDMGGEYYC++SDITCSFP NG FT+D Sbjct: 242 TCICCSGENAAVLHYGHAGAPNDRTIKDGDICLFDMGGEYYCFASDITCSFPANGKFTED 301 Query: 898 QKIVYEAVLAASRAVKNACKPGVRWTDMHLLAERIILSSLKEAGLLQGDVNEMMAVRLGA 1077 QK +YEAVL + R V + KPGV W DMH LA+RI L L GLL G V+ M+ V LGA Sbjct: 302 QKAIYEAVLRSCRTVMSTMKPGVWWPDMHRLADRIHLEELARIGLLSGSVDAMLQVHLGA 361 Query: 1078 VFMPHGLGHLLGVDVHDVGGYIEDAIPRSELSGLRSLRTTQLLQENMELTVEPGCYFIDI 1257 VFMPHGLGH LG+DVHDVGGY E + R + GLRSLRT + L+ M LTVEPG YFID Sbjct: 362 VFMPHGLGHFLGLDVHDVGGYPE-GVERIDEPGLRSLRTARHLEPGMVLTVEPGIYFIDH 420 Query: 1258 LLDAAINNPEQARFMVINEINRFTRFGGVRIEDDIIVTSTGIECMTEVPRTCEEIEQRMA 1437 LLD A+ +P QA F + RF FGGVRIE+D++VT +G+E +T VPRT EEIE MA Sbjct: 421 LLDQALADPAQACFFNQEVLQRFRNFGGVRIEEDVVVTDSGMELLTCVPRTVEEIEACMA 480 Query: 1438 GNDYMS 1455 G D S Sbjct: 481 GCDKAS 486
>sp|P43590|YFH6_YEAST Hypothetical 61.8 kDa peptidase in MPR1-GCN20 intergenic region Length = 535 Score = 213 bits (542), Expect = 1e-54 Identities = 139/424 (32%), Positives = 209/424 (49%), Gaps = 5/424 (1%) Frame = +1 Query: 172 DSALTFRQESYFHWAFGVLEPDFYGLIHIDSTKTILFMPRLDERYAIFMGALETKDDVKQ 351 D+ FRQ YF+ GV P L + + K LF+P +DE I+ G + D+ + Sbjct: 113 DTNKDFRQNRYFYHLSGVDIPASAILFNCSTDKLTLFLPNIDEEDVIWSGMPLSLDEAMR 172 Query: 352 KYAVDEVYYTDEILNVLEELKPSEIHTLHGLNTDSNNYTMEASFTGIANFQVINSVLHNE 531 + +DE Y ++ +EL+ I T N N + F ++ Sbjct: 173 VFDIDEALYISDLGKKFKELQDFAIFTTDLDNVHDENIARSLIPSDPNFFYAMD------ 226 Query: 532 ITECRVIKTNQELEVLRYANKASSKAHRDIMRSVHPGMYEYQIESLFLHHCYFHGAMRNV 711 E R IK E+E +R A + S K+H +M ++ + E QI++ F +H G R++ Sbjct: 227 --ETRAIKDWYEIESIRKACQISDKSHLAVMSALPIELNELQIQAEFEYHATRQGG-RSL 283 Query: 712 AYTSIGATGCNCSVLHYGHAGAPNDRKIEDGDMCLFDMGGEYYCYSSDITCSFPVNGHFT 891 Y I +G C LHY N I+ L D G E+ Y+SDIT FP +G FT Sbjct: 284 GYDPICCSGPACGTLHY----VKNSEDIKGKHSILIDAGAEWRQYTSDITRCFPTSGKFT 339 Query: 892 KDQKIVYEAVLAASRAVKNACKPGVRWTDMHLLAERIILSSLKEAGLLQGDV--NEMMAV 1065 + + VYE VL KPG +W D+H L ++++ G+ + + +E+ Sbjct: 340 AEHREVYETVLDMQNQAMERIKPGAKWDDLHALTHKVLIKHFLSMGIFKKEFSEDEIFKR 399 Query: 1066 RLGAVFMPHGLGHLLGVDVHDVGG--YIEDAIPRSELSGLRSLRTTQLLQENMELTVEPG 1239 R F PHGLGH+LG+DVHDVGG +D P R LR + L+ENM +T EPG Sbjct: 400 RASCAFYPHGLGHMLGLDVHDVGGNPNYDDPDPM-----FRYLRIRRPLKENMVITNEPG 454 Query: 1240 CYFIDILLDAAI-NNPEQARFMVINEINRFTRFGGVRIEDDIIVTSTGIECMTEVPRTCE 1416 CYF L+ + +PE+ + ++ + R+ GGVRIEDDI+VT G E +T + + Sbjct: 455 CYFNQFLIKEFLEKHPERLEVVDMSVLKRYMYVGGVRIEDDILVTKDGYENLTGITSDPD 514 Query: 1417 EIEQ 1428 EIE+ Sbjct: 515 EIEK 518
>sp|P15034|AMPP_ECOLI Xaa-Pro aminopeptidase (X-Pro aminopeptidase) (Aminopeptidase P II) (APP-II) (Aminoacylproline aminopeptidase) Length = 441 Score = 182 bits (462), Expect = 2e-45 Identities = 148/446 (33%), Positives = 215/446 (48%), Gaps = 24/446 (5%) Frame = +1 Query: 169 SDSALTFRQESYFHWAFGVLEPD-FYGLIHIDSTK--TILFMPRLDERYAIFMGALETKD 339 +DS +RQ S F + G EP+ LI D T ++LF D I+ G +D Sbjct: 38 ADSEYPYRQNSDFWYFTGFNEPEAVLVLIKSDDTHNHSVLFNRVRDLTAEIWFGRRLGQD 97 Query: 340 DVKQKYAVDEVYYTDEILNVLEELKPSEIHTLHGLNTDSNNYTMEASFTGIANFQVINSV 519 +K VD EI L +L L+GL+ + E ++ + ++NS Sbjct: 98 AAPEKLGVDRALAFSEINQQLYQL-------LNGLDVVYHAQG-EYAYADV----IVNSA 145 Query: 520 LHN--------------------EITECRVIKTNQELEVLRYANKASSKAHRDIMRSVHP 639 L + E R+ K+ +E+ VLR A + ++ AH M P Sbjct: 146 LEKLRKGSRQNLTAPATMIDWRPVVHEMRLFKSPEEIAVLRRAGEITAMAHTRAMEKCRP 205 Query: 640 GMYEYQIESLFLHHCYFHGAMRNVAYTSIGATGCNCSVLHYGHAGAPNDRKIEDGDMCLF 819 GM+EY +E H HGA R +Y +I +G N +LHY N+ ++ DGD+ L Sbjct: 206 GMFEYHLEGEIHHEFNRHGA-RYPSYNTIVGSGENGCILHY----TENECEMRDGDLVLI 260 Query: 820 DMGGEYYCYSSDITCSFPVNGHFTKDQKIVYEAVLAASRAVKNACKPGVRWTDMHLLAER 999 D G EY Y+ DIT +FPVNG FT+ Q+ +Y+ VL + +PG ++ R Sbjct: 261 DAGCEYKGYAGDITRTFPVNGKFTQAQREIYDIVLESLETSLRLYRPGTSILEVTGEVVR 320 Query: 1000 IILSSLKEAGLLQGDVNEMMAVRLGAVFMPHGLGHLLGVDVHDVGGYIEDAIPRSELSGL 1179 I++S L + G+L+GDV+E++A F HGL H LG+DVHDVG Y +D Sbjct: 321 IMVSGLVKLGILKGDVDELIAQNAHRPFFMHGLSHWLGLDVHDVGVYGQD---------- 370 Query: 1180 RSLRTTQLLQENMELTVEPGCYFIDILLDAAINNPEQARFMVINEINRFTRFGGVRIEDD 1359 +++L+ M LTVEPG Y I DA + PEQ R + G+RIEDD Sbjct: 371 ----RSRILEPGMVLTVEPGLY---IAPDAEV--PEQYRGI------------GIRIEDD 409 Query: 1360 IIVTSTGIECMT-EVPRTCEEIEQRM 1434 I++T TG E +T V + EEIE M Sbjct: 410 IVITETGNENLTASVVKKPEEIEALM 435
>sp|P44881|AMPP_HAEIN Xaa-Pro aminopeptidase (X-Pro aminopeptidase) (Aminopeptidase P II) (APP-II) (Aminoacylproline aminopeptidase) Length = 430 Score = 179 bits (454), Expect = 2e-44 Identities = 145/477 (30%), Positives = 230/477 (48%), Gaps = 7/477 (1%) Frame = +1 Query: 25 QLGDALSVPMTLHTENRKRLCAKMKATNKINNNAFILLQGGTDHYLGGSDSALTFRQESY 204 +L +P E R R+ A+M+ N+A +L +D FRQ+SY Sbjct: 2 ELAYMAKLPKEEFEERRTRVFAQMQP-----NSALLLFSEIEKRR--NNDCTYPFRQDSY 54 Query: 205 FHWAFGVLEPDFYGLIHIDST--KTILFMPRLDERYAIFMGALETKDDVKQKYAVDEVYY 378 F + G EP+ L+ K I+F+ D + G + Q+ V+E Y Sbjct: 55 FWYLTGFNEPNAALLLLKTEQVEKAIIFLRPRDPLLETWNGRRLGVERAPQQLNVNEAYS 114 Query: 379 TDEILNVLEELKPSEIHTLH--GLNTDSNNYTMEAS--FTGIANFQVINSVLHNEITECR 546 +E VL ++ + H ++T + E++ F+ I +++ + ++E R Sbjct: 115 IEEFATVLPKILKNLTALYHVPEIHTWGDTLVSESAVNFSEILDWRPM-------LSEMR 167 Query: 547 VIKTNQELEVLRYANKASSKAHRDIMRSVHPGMYEYQIESLFLHHCYFHGAMRNVAYTSI 726 +IK+ E+ +++ A + ++ H M++ P +EY+IES LH H A R +Y SI Sbjct: 168 LIKSPNEIRLMQQAGQITALGHIKAMQTTRPNRFEYEIESDILHEFNRHCA-RFPSYNSI 226 Query: 727 GATGCNCSVLHYGHAGAPNDRKIEDGDMCLFDMGGEYYCYSSDITCSFPVNGHFTKDQKI 906 A G N +LHY NDR + DGD+ L D G E+ Y+ DIT +FPVNG F++ Q+ Sbjct: 227 VAGGSNACILHY----TENDRPLNDGDLVLIDAGCEFAMYAGDITRTFPVNGKFSQPQRE 282 Query: 907 VYEAVLAASRAVKNACKPGVRWTDMHLLAERIILSSLKEAGLLQGDVNEMMAVRLGAVFM 1086 +YE VL A + PG + RI L + G+L+GDV+ ++ + F Sbjct: 283 IYELVLKAQKRAIELLVPGNSIKQANDEVIRIKTQGLVDLGILKGDVDTLIEQQAYRQFY 342 Query: 1087 PHGLGHLLGVDVHDVGGYIEDAIPRSELSGLRSLRTTQLLQENMELTVEPGCYFIDILLD 1266 HGLGH LG+DVHDVG Y +D ++L+ M +TVEPG Y + D Sbjct: 343 MHGLGHWLGLDVHDVGSYGQD--------------KQRILEIGMVITVEPGIYISE---D 385 Query: 1267 AAINNPEQARFMVINEINRFTRFGGVRIEDDIIVTSTGIECMT-EVPRTCEEIEQRM 1434 A + PEQ + + GVRIED++++T G + +T VP+ +IE M Sbjct: 386 ADV--PEQYKGI------------GVRIEDNLLMTEYGNKILTAAVPKEIADIENLM 428
>sp|P0A3Z2|AMPP1_STRLI Xaa-Pro aminopeptidase I (X-Pro aminopeptidase I) (Aminopeptidase P I) (APP) (PEPP I) (Aminoacylproline aminopeptidase I) sp|P0A3Z1|AMPP1_STRCO Xaa-Pro aminopeptidase I (X-Pro aminopeptidase I) (Aminopeptidase P I) (APP) (PEPP I) (Aminoacylproline aminopeptidase I) Length = 491 Score = 129 bits (325), Expect = 2e-29 Identities = 97/306 (31%), Positives = 154/306 (50%), Gaps = 4/306 (1%) Frame = +1 Query: 532 ITECRVIKTNQELEVLRYANKASSKAHRDIMRSVH--PGMYEYQIESLFLHHCYFHGAMR 705 ++E R++K E+ L+ A ++ + D+++ + E IE F G Sbjct: 216 LSEARLVKDEFEIGELQKAVDSTVRGFEDVVKVLDRAEATSERYIEGTFFLRARVEG--N 273 Query: 706 NVAYTSIGATGCNCSVLHYGHAGAPNDRKIEDGDMCLFDMGGEYYCY-SSDITCSFPVNG 882 +V Y SI A G + LH+ ND + GD+ L D G E + Y ++D+T + P++G Sbjct: 274 DVGYGSICAAGPHACTLHW----VRNDGPVRSGDLLLLDAGVETHTYYTADVTRTLPISG 329 Query: 883 HFTKDQKIVYEAVLAASRAVKNACKPGVRWTDMHLLAERIILSSLKEAGLLQGDVNEMMA 1062 +++ QK +Y+AV A A A +PG ++ D H ++R++ L E GL++G V ++ Sbjct: 330 TYSELQKKIYDAVYDAQEAGIAAVRPGAKYRDFHDASQRVLAERLVEWGLVEGPVERVLE 389 Query: 1063 VRLGAVFMPHGLGHLLGVDVHDVGGYIEDAIPRSELSGLRSLRTTQLLQENMELTVEPGC 1242 + L + HG GH+LG+DVHD A S + G L+ M LTVEPG Sbjct: 390 LGLQRRWTLHGTGHMLGMDVHDCAA----ARVESYVDG--------TLEPGMVLTVEPGL 437 Query: 1243 YFIDILLDAAINNPEQARFMVINEINRFTRFGGVRIEDDIIVTSTGIECMTE-VPRTCEE 1419 YF QA + + E R GVRIEDDI+VT+ G ++ +PR +E Sbjct: 438 YF-------------QADDLTVPEEYRGI---GVRIEDDILVTADGNRNLSAGLPRRSDE 481 Query: 1420 IEQRMA 1437 +E+ MA Sbjct: 482 VEEWMA 487
>sp|P0A3Z4|AMPP2_STRLI Xaa-Pro aminopeptidase II (X-Pro aminopeptidase II) (Aminopeptidase P II) (APP) (PEPP II) (Aminoacylproline aminopeptidase II) sp|P0A3Z3|AMPP2_STRCO Xaa-Pro aminopeptidase II (X-Pro aminopeptidase II) (Aminopeptidase P II) (APP) (PEPP II) (Aminoacylproline aminopeptidase II) Length = 470 Score = 120 bits (300), Expect = 1e-26 Identities = 97/310 (31%), Positives = 149/310 (48%), Gaps = 4/310 (1%) Frame = +1 Query: 520 LHNEITECRVIKTNQELEVLRYANKASSKAHRDIMRSVHPGMY--EYQIESLFLHHCYFH 693 L + +++ R++K EL LR A ++ + D++ + + E +E F Sbjct: 191 LEDALSDLRLVKDAWELGELRKAVDSTVRGFTDVVGELSRAVASSERWLEGTFFRRARLE 250 Query: 694 GAMRNVAYTSIGATGCNCSVLHYGHAGAPNDRKIEDGDMCLFDMGGEYYC-YSSDITCSF 870 G V Y +I A G + +++H+ ND + GD+ L D G E Y++D+T + Sbjct: 251 G--NAVGYGTICAAGEHATIMHW----TDNDGPVRPGDLLLLDAGVETRSLYTADVTRTL 304 Query: 871 PVNGHFTKDQKIVYEAVLAASRAVKNACKPGVRWTDMHLLAERIILSSLKEAGLLQGDVN 1050 P++G FT Q+ VY+AV A A KPG + D H A+R + + L E G ++G Sbjct: 305 PISGTFTPLQREVYDAVYEAQEAGIATVKPGAAYRDFHEAAQRHLAARLVEWGFIEGPAE 364 Query: 1051 EMMAVRLGAVFMPHGLGHLLGVDVHDVGGYIEDAIPRSELSGLRSLRTTQLLQENMELTV 1230 + L F G GH+LG+DVHD A R+E +L+ M LTV Sbjct: 365 RAYELGLQRRFTMAGTGHMLGLDVHDC------ARARTE------EYVEGVLEPGMCLTV 412 Query: 1231 EPGCYFIDILLDAAINNPEQARFMVINEINRFTRFGGVRIEDDIIVTSTGIECMTE-VPR 1407 EPG YF QA + + E R GVRIEDD++VT G E ++ +PR Sbjct: 413 EPGLYF-------------QADDLTVPEEWRGI---GVRIEDDLVVTEDGHENLSAGLPR 456 Query: 1408 TCEEIEQRMA 1437 + +E+E MA Sbjct: 457 SADEVEAWMA 466
>sp|P21165|PEPQ_ECOLI Xaa-Pro dipeptidase (X-Pro dipeptidase) (Proline dipeptidase) (Prolidase) (Imidodipeptidase) Length = 443 Score = 118 bits (295), Expect = 5e-26 Identities = 88/287 (30%), Positives = 137/287 (47%), Gaps = 3/287 (1%) Frame = +1 Query: 544 RVIKTNQELEVLRYANKASSKAHRDIMRSVHPGMYEYQIESLFLHHCYFHGAMRNVAYTS 723 R KT EL +R A K + HR + GM E+ I +L +V Y++ Sbjct: 159 RSFKTEYELACMREAQKMAVNGHRAAEEAFRSGMSEFDINIAYLTATGHRDT--DVPYSN 216 Query: 724 IGATGCNCSVLHYGHAGAPNDRKIEDGDMCLFDMGGEYYCYSSDITCSFPVNGHFTKDQK 903 I A + +VLHY + + E+ L D G EY Y++D+T ++ D Sbjct: 217 IVALNEHAAVLHYTKL---DHQAPEEMRSFLLDAGAEYNGYAADLTRTWSAKSD--NDYA 271 Query: 904 IVYEAVLAASRAVKNACKPGVRWTDMHLLAERIILSSLKEAGLLQGDVNEMMAVR-LGAV 1080 + + V A+ K GV + D H+ + I L++ ++ E M L Sbjct: 272 QLVKDVNDEQLALIATMKAGVSYVDYHIQFHQRIAKLLRKHQIITDMSEEAMVENDLTGP 331 Query: 1081 FMPHGLGHLLGVDVHDVGGYIED--AIPRSELSGLRSLRTTQLLQENMELTVEPGCYFID 1254 FMPHG+GH LG+ VHDV G+++D + + LR T++LQ M LT+EPG YFI+ Sbjct: 332 FMPHGIGHPLGLQVHDVAGFMQDDSGTHLAAPAKYPYLRCTRILQPGMVLTIEPGIYFIE 391 Query: 1255 ILLDAAINNPEQARFMVINEINRFTRFGGVRIEDDIIVTSTGIECMT 1395 LL A + ++ +I FGG+RIED++++ +E MT Sbjct: 392 SLL-APWREGQFSKHFNWQKIEALKPFGGIRIEDNVVIHENNVENMT 437
>sp|P81535|PEPQ_PYRFU Xaa-Pro dipeptidase (X-Pro dipeptidase) (Proline dipeptidase) (Prolidase) (Imidodipeptidase) Length = 348 Score = 108 bits (269), Expect = 5e-23 Identities = 103/388 (26%), Positives = 175/388 (45%) Frame = +1 Query: 244 GLIHIDSTKTILFMPRLDERYAIFMGALETKDDVKQKYAVDEVYYTDEILNVLEELKPSE 423 G I +D + L++P L+ M E+K V + DE+Y E LK +E Sbjct: 40 GYIIVDGDEATLYVPELEYE----MAKEESKLPVVKFKKFDEIY---------EILKNTE 86 Query: 424 IHTLHGLNTDSNNYTMEASFTGIANFQVINSVLHNEITECRVIKTNQELEVLRYANKASS 603 + G + +Y+M +F +N + + + I + R+IKT +E+E++ A + + Sbjct: 87 TLGIEG----TLSYSMVENFKEKSNVKEFKKI-DDVIKDLRIIKTKEEIEIIEKACEIAD 141 Query: 604 KAHRDIMRSVHPGMYEYQIESLFLHHCYFHGAMRNVAYTSIGATGCNCSVLHYGHAGAPN 783 KA + + G E ++ + + +GA + A+ +I A+G ++ H G + Sbjct: 142 KAVMAAIEEITEGKREREVAAKVEYLMKMNGAEKP-AFDTIIASGHRSALPH----GVAS 196 Query: 784 DRKIEDGDMCLFDMGGEYYCYSSDITCSFPVNGHFTKDQKIVYEAVLAASRAVKNACKPG 963 D++IE GD+ + D+G Y Y+SDIT + V G + Q+ +YE VL A + A KPG Sbjct: 197 DKRIERGDLVVIDLGALYNHYNSDITRTIVV-GSPNEKQREIYEIVLEAQKRAVEAAKPG 255 Query: 964 VRWTDMHLLAERIILSSLKEAGLLQGDVNEMMAVRLGAVFMPHGLGHLLGVDVHDVGGYI 1143 + ++ +A II KE G GD + H LGH +G+++H+ Sbjct: 256 MTAKELDSIAREII----KEYG--YGD------------YFIHSLGHGVGLEIHE----- 292 Query: 1144 EDAIPRSELSGLRSLRTTQLLQENMELTVEPGCYFIDILLDAAINNPEQARFMVINEINR 1323 PR S +L+E M +T+EPG Y Sbjct: 293 ---WPRI------SQYDETVLKEGMVITIEPGIY-------------------------- 317 Query: 1324 FTRFGGVRIEDDIIVTSTGIECMTEVPR 1407 + GGVRIED +++T G + +T+ R Sbjct: 318 IPKLGGVRIEDTVLITENGAKRLTKTER 345
>sp|P40051|YEQ8_YEAST Hypothetical 58.0 kDa peptidase in PTP3-ILV1 intergenic region Length = 511 Score = 101 bits (251), Expect = 7e-21 Identities = 118/457 (25%), Positives = 191/457 (41%), Gaps = 34/457 (7%) Frame = +1 Query: 133 LLQGGTDHYLGGSDSALTFRQESYFHWAFGVLEPDFYGLIHIDSTKTILFMPRLDERYAI 312 ++ G D F+QE+ + G EP+ + IL P I Sbjct: 87 VILAGNDIQFASGAVFYPFQQENDLFYLSGWNEPN---------SVMILEKPTDSLSDTI 137 Query: 313 FMGALETKDDVKQK--------YAVDEVYYTDEI--LNVLEELKPSEIHT--------LH 438 F + KD +K Y V E++ DE +N L + P I+ L Sbjct: 138 FHMLVPPKDAFAEKWEGFRSGVYGVQEIFNADESASINDLSKYLPKIINRNDFIYFDMLS 197 Query: 439 GLNTDSNNYT-MEASFTGIANF-QVINSV-------LHNEITECRVIKTNQELEVLRYAN 591 N S+N+ +++ G N + +NS+ + I E R IK+ QEL ++R A Sbjct: 198 TSNPSSSNFKHIKSLLDGSGNSNRSLNSIANKTIKPISKRIAEFRKIKSPQELRIMRRAG 257 Query: 592 KASSKAHRDIMRSVHPGMYEYQIESLFLHHCYFHGAMRNVAYTSIGATGCNCSVLHYGHA 771 + S ++ E ++S FLH+ + G AY + ATG N +HY Sbjct: 258 QISGRSFNQAFAKRFRN--ERTLDS-FLHYKFISGGCDKDAYIPVVATGSNSLCIHYTR- 313 Query: 772 GAPNDRKIEDGDMCLFDMGGEYYCYSSDITCSFPVNGHFTKDQKIVYEAVLAASRAVKNA 951 ND + D +M L D G Y +DI+ ++P +G FT Q+ +YEAVL R Sbjct: 314 ---NDDVMFDDEMVLVDAAGSLGGYCADISRTWPNSGKFTDAQRDLYEAVLNVQRDCIKL 370 Query: 952 CKPGVRWT--DMHLLAERIILSSLKEAGLLQGDVNEMMAVRLGAVFMPHGLGHLLGVDVH 1125 CK ++ D+H + ++ LK G ++++ + ++ PH +GH LG+DVH Sbjct: 371 CKASNNYSLHDIHEKSITLMKQELKNLG-----IDKVSGWNVEKLY-PHYIGHNLGLDVH 424 Query: 1126 DVGGYIEDAIPRSELSGLRSLRTTQLLQENMELTVEPGCYFIDILLDAAINNPEQARFMV 1305 DV ++S L+ Q+ +T+EPG Y P + F Sbjct: 425 DV----------PKVSRYEPLKVGQV------ITIEPGLYI-----------PNEESF-- 455 Query: 1306 INEINRFTRFGGVRIEDDIIV-----TSTGIECMTEV 1401 + R G+RIEDDI + T+ +E + E+ Sbjct: 456 ----PSYFRNVGIRIEDDIAIGEDTYTNLTVEAVKEI 488
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 165,047,772
Number of Sequences: 369166
Number of extensions: 3389040
Number of successful extensions: 8386
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 7893
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 8306
length of database: 68,354,980
effective HSP length: 115
effective length of database: 47,110,455
effective search space used: 18467298360
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail