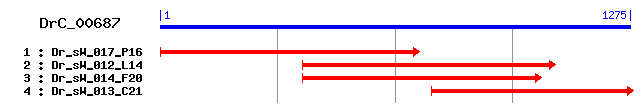
DrC_00687
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00687 (1275 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P13017|AMPE_ECOLI Protein ampE 34 1.0 sp|P07661|CIT1_ECOLI Citrate-proton symporter (Citrate tran... 33 1.8 sp|P29086|YSO2_ACIAM Hypothetical transport protein in sor ... 32 3.9 sp|P60363|HPPA1_RHOPA Pyrophosphate-energized proton pump (... 31 8.8
>sp|P13017|AMPE_ECOLI Protein ampE Length = 284 Score = 33.9 bits (76), Expect = 1.0 Identities = 36/129 (27%), Positives = 53/129 (41%), Gaps = 12/129 (9%) Frame = +1 Query: 712 TLMGTAILTCFGYWL------------GVSAIMWVLAYVGNTKLRPVQINTVMGYALTGH 855 TLMG A L + YWL G+ A++ VL +V PV++ V+ YAL GH Sbjct: 161 TLMGYAFLRAWQYWLARYQTPHHRLQSGIDAVLHVLDWV------PVRLAGVV-YALIGH 213 Query: 856 CIVVLLGSLIHFGNSHVPFYIVWVICGGLCSARVVTIFWARTPGHSERIISAITIVVLHM 1035 L G+ H Y V AR + TP + + + VV+ + Sbjct: 214 GEKALPAWFASLGDFHTSQYQVLTRLAQFSLAREPHVDKVETPKAAVSMAKKTSFVVVVV 273 Query: 1036 GFLLYLHFA 1062 LL ++ A Sbjct: 274 IALLTIYGA 282
>sp|P07661|CIT1_ECOLI Citrate-proton symporter (Citrate transporter) (Citrate carrier protein) (Citrate utilization determinant) (Citrate utilization protein A) Length = 431 Score = 33.1 bits (74), Expect = 1.8 Identities = 33/125 (26%), Positives = 56/125 (44%), Gaps = 10/125 (8%) Frame = +1 Query: 634 PVMICLTLIALLL-FEMKTSGTKVQEGTLMGTAILTCFGYWLGVSAIMWVLAYVGNTKLR 810 PV++ +TL+AL+ + T + T M T +L F ++ G+ V A T++ Sbjct: 302 PVLMGITLLALVTTLPVMNWLTAAPDFTRM-TLVLLWFSFFFGMYNGAMVAAL---TEVM 357 Query: 811 PVQINTVMGYALTGHCIVVLLGSLIHF---------GNSHVPFYIVWVICGGLCSARVVT 963 PV + TV G++L + G L G+ P + W++C LC T Sbjct: 358 PVYVRTV-GFSLAFSLATAIFGGLTPAISTALVQLTGDKSSPGW--WLMCAALCGLAATT 414 Query: 964 IFWAR 978 + +AR Sbjct: 415 MLFAR 419
>sp|P29086|YSO2_ACIAM Hypothetical transport protein in sor 3'region (ORF2) Length = 253 Score = 32.0 bits (71), Expect = 3.9 Identities = 34/113 (30%), Positives = 51/113 (45%), Gaps = 12/113 (10%) Frame = +1 Query: 628 YGPVMICLTLIALLLFEMKTSGTKVQEGTLMGTAILTCFGYWLGV------------SAI 771 Y +I + L L+L E KT+ T+V G L+G + + + LG+ +A Sbjct: 59 YTMPLIAIPLSTLILRE-KTTKTEVI-GILIGFSGVVIYSLNLGIYFSLIGIVLTLINAF 116 Query: 772 MWVLAYVGNTKLRPVQINTVMGYALTGHCIVVLLGSLIHFGNSHVPFYIVWVI 930 W L V KLR +V + + +LLGSLI F S + FY + I Sbjct: 117 FWALFTVYFRKLRGFDATSV-------NAVQLLLGSLIFFTLSPIQFYFKYSI 162
>sp|P60363|HPPA1_RHOPA Pyrophosphate-energized proton pump (Pyrophosphate-energized inorganic pyrophosphatase) (H+-PPase) (Membrane-bound proton-translocating pyrophosphatase) Length = 706 Score = 30.8 bits (68), Expect = 8.8 Identities = 23/84 (27%), Positives = 41/84 (48%), Gaps = 1/84 (1%) Frame = +1 Query: 721 GTAILTCFGYWLGVSA-IMWVLAYVGNTKLRPVQINTVMGYALTGHCIVVLLGSLIHFGN 897 G A+ C L V+ I+W+ Y T RPV+ ++ ++TGH V+ G I + Sbjct: 330 GLALFECGIVGLAVTGLIIWITEYYTGTDFRPVK--SIAQASVTGHGTNVIQGLAISMES 387 Query: 898 SHVPFYIVWVICGGLCSARVVTIF 969 + +P + +I G L + + +F Sbjct: 388 TALPAIV--IIAGILITYSLAGLF 409
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 126,029,202
Number of Sequences: 369166
Number of extensions: 2452458
Number of successful extensions: 5193
Number of sequences better than 10.0: 4
Number of HSP's better than 10.0 without gapping: 5058
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 5193
length of database: 68,354,980
effective HSP length: 113
effective length of database: 47,479,925
effective search space used: 14766256675
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail