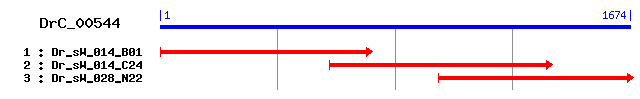
DrC_00544
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00544 (1674 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|P22033|MUTA_HUMAN Methylmalonyl-CoA mutase, mitochondria... 733 0.0 sp|Q8HXX1|MUTA_MACFA Methylmalonyl-CoA mutase, mitochondria... 732 0.0 sp|Q8MI68|MUTA_PIG Methylmalonyl-CoA mutase, mitochondrial ... 722 0.0 sp|Q23381|MUTA_CAEEL Probable methylmalonyl-CoA mutase, mit... 717 0.0 sp|P16332|MUTA_MOUSE Methylmalonyl-CoA mutase, mitochondria... 702 0.0 sp|Q59677|MUTB_PORGI Methylmalonyl-CoA mutase large subunit... 613 e-175 sp|Q05065|MUTB_STRCM Methylmalonyl-CoA mutase large subunit... 590 e-168 sp|O86028|MUTB_RHIME Methylmalonyl-CoA mutase (MCM) 567 e-161 sp|P65487|MUTB_MYCTU Probable methylmalonyl-CoA mutase larg... 566 e-161 sp|P11653|MUTB_PROFR Methylmalonyl-CoA mutase large subunit... 566 e-161
>sp|P22033|MUTA_HUMAN Methylmalonyl-CoA mutase, mitochondrial precursor (MCM) (Methylmalonyl-CoA isomerase) Length = 750 Score = 733 bits (1892), Expect = 0.0 Identities = 361/503 (71%), Positives = 415/503 (82%) Frame = +3 Query: 3 PSMKIIGDIFEYTAKHMPKFNSISISGYHMQEAGANAILEMAYTIADGLEYCRTGLKAGL 182 PSMKII DIFEYTAKHMPKFNSISISGYHMQEAGA+AILE+AYT+ADGLEY RTGL+AGL Sbjct: 237 PSMKIIADIFEYTAKHMPKFNSISISGYHMQEAGADAILELAYTLADGLEYSRTGLQAGL 296 Query: 183 HIDEFAPRLSFFWGVGMNFYMEIAKMRAARRVWAELLKSKFNSKDKKSLLLRAHSQTSGW 362 IDEFAPRLSFFWG+GMNFYMEIAKMRA RR+WA L++ F K+ KSLLLRAH QTSGW Sbjct: 297 TIDEFAPRLSFFWGIGMNFYMEIAKMRAGRRLWAHLIEKMFQPKNSKSLLLRAHCQTSGW 356 Query: 363 SLTEQDPYNNIMRTTIEAMAAVFGGTQSLHTNAFDEALGLPTKFSARIARNXXXXXXXXX 542 SLTEQDPYNNI+RT IEAMAAVFGGTQSLHTN+FDEALGLPT SARIARN Sbjct: 357 SLTEQDPYNNIVRTAIEAMAAVFGGTQSLHTNSFDEALGLPTVKSARIARNTQIIIQEES 416 Query: 543 XXXKVADPWAGSFMMESLTQEIYSEAIKVIEEIEAMGGMAKAVSEGIPKLKIEECAAKRQ 722 KVADPW GS+MME LT ++Y A+K+I EIE MGGMAKAV+EGIPKL+IEECAA+RQ Sbjct: 417 GIPKVADPWGGSYMMECLTNDVYDAALKLINEIEEMGGMAKAVAEGIPKLRIEECAARRQ 476 Query: 723 AHIDSGQEVIVGVNKYRLENPEKVDVLHIDNTKVRESQIAKINALKQSRDNNKAQNILKK 902 A IDSG EVIVGVNKY+LE + V+VL IDNT VR QI K+ +K SRD A+ L Sbjct: 477 ARIDSGSEVIVGVNKYQLEKEDAVEVLAIDNTSVRNRQIEKLKKIKSSRDQALAERCLAA 536 Query: 903 LEECCRSGHGNLLEIAVDASRARCTVGEITDAMVAAYGRHVAVGRLVSGAYKSEFGDNKE 1082 L EC SG GN+L +AVDASRARCTVGEITDA+ +G H A R+VSGAY+ EFG++KE Sbjct: 537 LTECAASGDGNILALAVDASRARCTVGEITDALKKVFGEHKANDRMVSGAYRQEFGESKE 596 Query: 1083 IAAVLKKVEDFLKNEGRRPRILIAKLGQDGHDRGGKVIATGFSDLGFDVDVGPLFSTPKE 1262 I + +K+V F++ EGRRPR+L+AK+GQDGHDRG KVIATGF+DLGFDVD+GPLF TP+E Sbjct: 597 ITSAIKRVHKFMEREGRRPRLLVAKMGQDGHDRGAKVIATGFADLGFDVDIGPLFQTpre 656 Query: 1263 AVEQAVDADVHVIGVSSLAAGHMTLVPQLIDELKKAGRSDIVVVAGGVIPPQDYEFLRKV 1442 +QAVDADVH +GVS+LAAGH TLVP+LI EL GR DI+V+ GGVIPPQDYEFL +V Sbjct: 657 VAQQAVDADVHAVGVSTLAAGHKTLVPELIKELNSLGRPDILVMCGGVIPPQDYEFLFEV 716 Query: 1443 GVSEIFGPGTRIPDAASKVVDAI 1511 GVS +FGPGTRIP AA +V+D I Sbjct: 717 GVSNVFGPGTRIPKAAVQVLDDI 739
>sp|Q8HXX1|MUTA_MACFA Methylmalonyl-CoA mutase, mitochondrial precursor (MCM) (Methylmalonyl-CoA isomerase) Length = 750 Score = 732 bits (1890), Expect = 0.0 Identities = 360/503 (71%), Positives = 415/503 (82%) Frame = +3 Query: 3 PSMKIIGDIFEYTAKHMPKFNSISISGYHMQEAGANAILEMAYTIADGLEYCRTGLKAGL 182 PSMKII DIF+YT+KHMPKFNSISISGYHMQEAGA+AILE+AYT+ADGLEY RTGL+AGL Sbjct: 237 PSMKIIADIFQYTSKHMPKFNSISISGYHMQEAGADAILELAYTLADGLEYSRTGLQAGL 296 Query: 183 HIDEFAPRLSFFWGVGMNFYMEIAKMRAARRVWAELLKSKFNSKDKKSLLLRAHSQTSGW 362 IDEFAPRLSFFWG+GMNFYMEIAKMRA RR+WA L++ F K KSLLLRAH QTSGW Sbjct: 297 TIDEFAPRLSFFWGIGMNFYMEIAKMRAGRRLWAHLIEKMFQPKSSKSLLLRAHCQTSGW 356 Query: 363 SLTEQDPYNNIMRTTIEAMAAVFGGTQSLHTNAFDEALGLPTKFSARIARNXXXXXXXXX 542 SLTEQDPYNNI+RT IEAMAAVFGGTQSLHTN+FDEALGLPT SARIARN Sbjct: 357 SLTEQDPYNNIVRTAIEAMAAVFGGTQSLHTNSFDEALGLPTVKSARIARNTQIIIQEES 416 Query: 543 XXXKVADPWAGSFMMESLTQEIYSEAIKVIEEIEAMGGMAKAVSEGIPKLKIEECAAKRQ 722 KVADPW GS+MMESLT ++Y A+K+I EIE MGGMAKAV+EGIPKL+IEECAA+RQ Sbjct: 417 GIPKVADPWGGSYMMESLTNDVYDAALKLINEIEEMGGMAKAVAEGIPKLRIEECAARRQ 476 Query: 723 AHIDSGQEVIVGVNKYRLENPEKVDVLHIDNTKVRESQIAKINALKQSRDNNKAQNILKK 902 A IDSG EVIVGVNKY+LE + V+VL IDNT VR QI K+ +K SRD A+ L Sbjct: 477 ARIDSGSEVIVGVNKYQLEKEDAVEVLAIDNTSVRNRQIEKLQKIKSSRDQALAERCLAA 536 Query: 903 LEECCRSGHGNLLEIAVDASRARCTVGEITDAMVAAYGRHVAVGRLVSGAYKSEFGDNKE 1082 L EC SG GN+L +AVDASRARCTVGEITDA+ +G H A R+VSGAY+ EFG++KE Sbjct: 537 LTECAASGDGNILALAVDASRARCTVGEITDALKKVFGEHKANDRMVSGAYRQEFGESKE 596 Query: 1083 IAAVLKKVEDFLKNEGRRPRILIAKLGQDGHDRGGKVIATGFSDLGFDVDVGPLFSTPKE 1262 I + +K+V F++ EGRRPR+L+AK+GQDGHDRG KVIATGF+DLGFDVD+GPLF TP+E Sbjct: 597 ITSAIKRVHKFMEREGRRPRLLVAKMGQDGHDRGAKVIATGFADLGFDVDIGPLFQTpre 656 Query: 1263 AVEQAVDADVHVIGVSSLAAGHMTLVPQLIDELKKAGRSDIVVVAGGVIPPQDYEFLRKV 1442 +QAVDADVH +GVS+LAAGH TLVP+LI EL GR DI+V+ GGVIPPQDYEFL +V Sbjct: 657 VAQQAVDADVHAVGVSTLAAGHKTLVPELIKELNSLGRPDILVMCGGVIPPQDYEFLFEV 716 Query: 1443 GVSEIFGPGTRIPDAASKVVDAI 1511 GVS +FGPGTRIP AA +V+D I Sbjct: 717 GVSNVFGPGTRIPKAAVQVLDDI 739
>sp|Q8MI68|MUTA_PIG Methylmalonyl-CoA mutase, mitochondrial precursor (MCM) (Methylmalonyl-CoA isomerase) Length = 750 Score = 722 bits (1864), Expect = 0.0 Identities = 356/503 (70%), Positives = 413/503 (82%) Frame = +3 Query: 3 PSMKIIGDIFEYTAKHMPKFNSISISGYHMQEAGANAILEMAYTIADGLEYCRTGLKAGL 182 PSMKII DIF+YTAKHMPKFNSISISGYHMQEAGA+AILE+AYTIADGLEYCRTGL+AGL Sbjct: 237 PSMKIIADIFQYTAKHMPKFNSISISGYHMQEAGADAILELAYTIADGLEYCRTGLQAGL 296 Query: 183 HIDEFAPRLSFFWGVGMNFYMEIAKMRAARRVWAELLKSKFNSKDKKSLLLRAHSQTSGW 362 IDEFAPRLSFFWG+GMNFYMEIAKMRA RR+WA L++ F K+ KSLLLRAH QTSGW Sbjct: 297 TIDEFAPRLSFFWGIGMNFYMEIAKMRAGRRLWAHLIEKMFRRKNSKSLLLRAHCQTSGW 356 Query: 363 SLTEQDPYNNIMRTTIEAMAAVFGGTQSLHTNAFDEALGLPTKFSARIARNXXXXXXXXX 542 SLTEQDPYNNI+RTT+EAMAAVFGGTQSLHTN+FDEALGLPT SARIARN Sbjct: 357 SLTEQDPYNNIIRTTVEAMAAVFGGTQSLHTNSFDEALGLPTVKSARIARNTQIIIQEES 416 Query: 543 XXXKVADPWAGSFMMESLTQEIYSEAIKVIEEIEAMGGMAKAVSEGIPKLKIEECAAKRQ 722 KVADPW GS+MMESLT ++Y A+K+I EIE MGGMAKAV+EGIPKL+IEECAA+RQ Sbjct: 417 GIPKVADPWGGSYMMESLTNDVYDAALKLINEIEEMGGMAKAVAEGIPKLRIEECAARRQ 476 Query: 723 AHIDSGQEVIVGVNKYRLENPEKVDVLHIDNTKVRESQIAKINALKQSRDNNKAQNILKK 902 A IDSG EVIVGVNKY+LE E V+VL IDNT VR QI K+ + + + L Sbjct: 477 ARIDSGSEVIVGVNKYQLEKEESVEVLAIDNTSVRNKQIEKLKKVNPAGIKLWLERCLTA 536 Query: 903 LEECCRSGHGNLLEIAVDASRARCTVGEITDAMVAAYGRHVAVGRLVSGAYKSEFGDNKE 1082 L C SG GN+L +AV+A+RARCTVGEITDAM +G H A R+VSGAY+ EFG++KE Sbjct: 537 LTACAASGDGNILALAVEATRARCTVGEITDAMKKVFGEHKANDRMVSGAYRQEFGESKE 596 Query: 1083 IAAVLKKVEDFLKNEGRRPRILIAKLGQDGHDRGGKVIATGFSDLGFDVDVGPLFSTPKE 1262 I+ +K+V F++ EGRRPR+L+AK+GQDGHDRG KVIATGF+DLGFDVD+GPLF TP+E Sbjct: 597 ISFAIKRVHKFMEREGRRPRLLVAKMGQDGHDRGAKVIATGFADLGFDVDIGPLFQTpre 656 Query: 1263 AVEQAVDADVHVIGVSSLAAGHMTLVPQLIDELKKAGRSDIVVVAGGVIPPQDYEFLRKV 1442 +QAVDADVH +GVS+LAAGH TLVP+LI EL GR DI+V+ GGVIPPQDYEFL +V Sbjct: 657 VAQQAVDADVHAVGVSTLAAGHKTLVPELIKELSTLGRPDILVMCGGVIPPQDYEFLFEV 716 Query: 1443 GVSEIFGPGTRIPDAASKVVDAI 1511 GVS +FGPGTRIP AA +V++ I Sbjct: 717 GVSNVFGPGTRIPKAAVQVLNDI 739
>sp|Q23381|MUTA_CAEEL Probable methylmalonyl-CoA mutase, mitochondrial precursor (MCM) Length = 744 Score = 717 bits (1851), Expect = 0.0 Identities = 351/512 (68%), Positives = 426/512 (83%) Frame = +3 Query: 3 PSMKIIGDIFEYTAKHMPKFNSISISGYHMQEAGANAILEMAYTIADGLEYCRTGLKAGL 182 PSM+IIGDIF YT++ MPKFNSISISGYHMQEAGA+A+LEMA+TIADG++YC TGL AGL Sbjct: 229 PSMRIIGDIFAYTSREMPKFNSISISGYHMQEAGADAVLEMAFTIADGIQYCETGLNAGL 288 Query: 183 HIDEFAPRLSFFWGVGMNFYMEIAKMRAARRVWAELLKSKFNSKDKKSLLLRAHSQTSGW 362 ID FAPRLSFFWG+ MNFYMEIAKMRAARR+WA L+K +F+ K KS++LR HSQTSGW Sbjct: 289 TIDAFAPRLSFFWGISMNFYMEIAKMRAARRLWANLIKERFSPKSDKSMMLRTHSQTSGW 348 Query: 363 SLTEQDPYNNIMRTTIEAMAAVFGGTQSLHTNAFDEALGLPTKFSARIARNXXXXXXXXX 542 SLTEQDPYNNI+RTTIEAMA+VFGGTQSLHTN+FDEALGLPTKFSARIARN Sbjct: 349 SLTEQDPYNNIIRTTIEAMASVFGGTQSLHTNSFDEALGLPTKFSARIARNTQIIIQEES 408 Query: 543 XXXKVADPWAGSFMMESLTQEIYSEAIKVIEEIEAMGGMAKAVSEGIPKLKIEECAAKRQ 722 VADPW GS+MMESLT EIY +A+ VI+EI+ +GGMAKAV+ G+ KLKIEE AAK+Q Sbjct: 409 GICNVADPWGGSYMMESLTDEIYEKALAVIKEIDELGGMAKAVASGMTKLKIEEAAAKKQ 468 Query: 723 AHIDSGQEVIVGVNKYRLENPEKVDVLHIDNTKVRESQIAKINALKQSRDNNKAQNILKK 902 A ID+G++VIVGVNKYRL++ ++V+VL IDN KVRE Q AK+N ++ +RD KAQ L Sbjct: 469 ARIDAGKDVIVGVNKYRLDHEQQVEVLKIDNAKVREEQCAKLNHIRATRDAEKAQKALDA 528 Query: 903 LEECCRSGHGNLLEIAVDASRARCTVGEITDAMVAAYGRHVAVGRLVSGAYKSEFGDNKE 1082 + E R G+GNL+E+A++A+RARCTVGEI+DAM + RH AV RLVSGAYKSEFG+ E Sbjct: 529 ITEGAR-GNGNLMELAIEAARARCTVGEISDAMEKVFNRHAAVNRLVSGAYKSEFGETSE 587 Query: 1083 IAAVLKKVEDFLKNEGRRPRILIAKLGQDGHDRGGKVIATGFSDLGFDVDVGPLFSTPKE 1262 ++ VL++V+ F +GR+PRI++AK+GQDGHDRG KVIATGF+DLGFDVDVGPLF TP E Sbjct: 588 MSQVLERVKSFADRDGRQPRIMVAKMGQDGHDRGAKVIATGFADLGFDVDVGPLFQTPLE 647 Query: 1263 AVEQAVDADVHVIGVSSLAAGHMTLVPQLIDELKKAGRSDIVVVAGGVIPPQDYEFLRKV 1442 A +QAVDADVHVIG SSLAAGH+TL+PQLI ELKK GR DI+VVAGGVIPPQDY+ L Sbjct: 648 AAQQAVDADVHVIGASSLAAGHLTLIPQLIGELKKLGRPDILVVAGGVIPPQDYKELYDA 707 Query: 1443 GVSEIFGPGTRIPDAASKVVDAIISNQNMAGG 1538 GV+ +FGPGTR+P A+++++ + +N A G Sbjct: 708 GVALVFGPGTRLPACANQILEKLEANLPEAPG 739
>sp|P16332|MUTA_MOUSE Methylmalonyl-CoA mutase, mitochondrial precursor (MCM) (Methylmalonyl-CoA isomerase) Length = 748 Score = 702 bits (1812), Expect = 0.0 Identities = 346/503 (68%), Positives = 405/503 (80%) Frame = +3 Query: 3 PSMKIIGDIFEYTAKHMPKFNSISISGYHMQEAGANAILEMAYTIADGLEYCRTGLKAGL 182 PSMKII DIF+YTA+HMPKFN ISISGYHMQEAG +AILE+AYTIADGLEYCRTGL+AGL Sbjct: 235 PSMKIIADIFQYTAQHMPKFNPISISGYHMQEAGTDAILELAYTIADGLEYCRTGLQAGL 294 Query: 183 HIDEFAPRLSFFWGVGMNFYMEIAKMRAARRVWAELLKSKFNSKDKKSLLLRAHSQTSGW 362 IDEFAPRLSFFWG+GMNFYMEIAKMRA RR+WA L++ F K+ KSLLLRAH QTSGW Sbjct: 295 TIDEFAPRLSFFWGIGMNFYMEIAKMRAGRRLWAHLIEKMFQPKNSKSLLLRAHCQTSGW 354 Query: 363 SLTEQDPYNNIMRTTIEAMAAVFGGTQSLHTNAFDEALGLPTKFSARIARNXXXXXXXXX 542 SLTEQDPYNNI+RT IEAMAAVFGGTQSLH+N+FDEALGLPT SARIARN Sbjct: 355 SLTEQDPYNNIVRTAIEAMAAVFGGTQSLHSNSFDEALGLPTVKSARIARNTRIIIQEES 414 Query: 543 XXXKVADPWAGSFMMESLTQEIYSEAIKVIEEIEAMGGMAKAVSEGIPKLKIEECAAKRQ 722 KVADPW GS+MMESLT ++Y A+K+I E+E MGGMAKAV+EGIPKL+IEECAA+RQ Sbjct: 415 GIPKVADPWGGSYMMESLTNDVYEAALKLIYEVEEMGGMAKAVAEGIPKLRIEECAARRQ 474 Query: 723 AHIDSGQEVIVGVNKYRLENPEKVDVLHIDNTKVRESQIAKINALKQSRDNNKAQNILKK 902 A IDSG EVIVGVNKY LE + V +L ID +R+ QI K+ +K SRD A+ L Sbjct: 475 ARIDSGSEVIVGVNKYHLEKEDSVHLLAIDIISLRKKQIEKLKKIKSSRDQALAEQCLSA 534 Query: 903 LEECCRSGHGNLLEIAVDASRARCTVGEITDAMVAAYGRHVAVGRLVSGAYKSEFGDNKE 1082 L +C SG GN+L +AVDA+RARCTV EITDA +G H A R+VSGAY+ EFG++KE Sbjct: 535 LTQCAASGDGNILALAVDAARARCTVPEITDAFKKVFGEHKANDRMVSGAYRQEFGESKE 594 Query: 1083 IAAVLKKVEDFLKNEGRRPRILIAKLGQDGHDRGGKVIATGFSDLGFDVDVGPLFSTPKE 1262 I + +K+V F++ EGRR +L+AK+G+DGHDRG KVIATGF+DLGFDVD+GPLF TP+E Sbjct: 595 ITSAIKRVNKFMEREGRRLGLLVAKMGKDGHDRGAKVIATGFADLGFDVDIGPLFQTpre 654 Query: 1263 AVEQAVDADVHVIGVSSLAAGHMTLVPQLIDELKKAGRSDIVVVAGGVIPPQDYEFLRKV 1442 AVDADVH +GVS+ AAGH TLVP+LI EL GR DI+V+ GGVIPPQDYEFL +V Sbjct: 655 VAHDAVDADVHAVGVSTHAAGHKTLVPELIKELTALGRPDILVMCGGVIPPQDYEFLYEV 714 Query: 1443 GVSEIFGPGTRIPDAASKVVDAI 1511 GVS +FGPGTRIP AA +V+D I Sbjct: 715 GVSNVFGPGTRIPRAAVQVLDDI 737
>sp|Q59677|MUTB_PORGI Methylmalonyl-CoA mutase large subunit (MCM-alpha) Length = 715 Score = 613 bits (1580), Expect = e-175 Identities = 301/503 (59%), Positives = 385/503 (76%) Frame = +3 Query: 6 SMKIIGDIFEYTAKHMPKFNSISISGYHMQEAGANAILEMAYTIADGLEYCRTGLKAGLH 185 SM+II DIFEYT+++MPKFNSISISGYHMQEAGA A +EMAYT+ADG++Y + G+ AG+ Sbjct: 212 SMRIIADIFEYTSQNMPKFNSISISGYHMQEAGATADIEMAYTLADGMQYLKAGIDAGID 271 Query: 186 IDEFAPRLSFFWGVGMNFYMEIAKMRAARRVWAELLKSKFNSKDKKSLLLRAHSQTSGWS 365 +D FAPRLSFFW +G+N +MEIAKMRAAR +WA+++KS F +K+ KSL LR HSQTSGWS Sbjct: 272 VDAFAPRLSFFWAIGVNHFMEIAKMRAARLLWAKIVKS-FGAKNPKSLALRTHSQTSGWS 330 Query: 366 LTEQDPYNNIMRTTIEAMAAVFGGTQSLHTNAFDEALGLPTKFSARIARNXXXXXXXXXX 545 LTEQDP+NN+ RT IEAMAA G TQSLHTNA DEA+ LPT FSARIARN Sbjct: 331 LTEQDPFNNVGRTCIEAMAAALGHTQSLHTNALDEAIALPTDFSARIARNTQIYIQEETL 390 Query: 546 XXKVADPWAGSFMMESLTQEIYSEAIKVIEEIEAMGGMAKAVSEGIPKLKIEECAAKRQA 725 K DPW GS+ +ESLT E+ +A +I+E++ MGGMAKA+ G+PKL+IEE AA+ QA Sbjct: 391 VCKEIDPWGGSYYVESLTNELVHKAWTLIKEVQEMGGMAKAIETGLPKLRIEEAAARTQA 450 Query: 726 HIDSGQEVIVGVNKYRLENPEKVDVLHIDNTKVRESQIAKINALKQSRDNNKAQNILKKL 905 IDS Q+VIVGVNKYRL + +D+L IDNT VR+ QI ++N L+ RD Q L+ + Sbjct: 451 RIDSHQQVIVGVNKYRLPKEDPIDILEIDNTAVRKQQIERLNDLRSHRDEKAVQEALEAI 510 Query: 906 EECCRSGHGNLLEIAVDASRARCTVGEITDAMVAAYGRHVAVGRLVSGAYKSEFGDNKEI 1085 +C + GNLL++AV A+ R ++GEI+DA GR+ AV R +SG Y SE G++K+ Sbjct: 511 TKCVETKEGNLLDLAVKAAGLRASLGEISDACEKVVGRYKAVIRTISGVYSSESGEDKDF 570 Query: 1086 AAVLKKVEDFLKNEGRRPRILIAKLGQDGHDRGGKVIATGFSDLGFDVDVGPLFSTPKEA 1265 A + E F K EGR+PRI+IAK+GQDGHDRG KV+ATG++D GFDVD+GPLF TP+EA Sbjct: 571 AHAKELAEKFAKKEGRQPRIMIAKMGQDGHDRGAKVVATGYADCGFDVDMGPLFQTPEEA 630 Query: 1266 VEQAVDADVHVIGVSSLAAGHMTLVPQLIDELKKAGRSDIVVVAGGVIPPQDYEFLRKVG 1445 QAV+ DVHV+GVSSLAAGH TL+PQ+I EL+K GR DI+V AGGVIP QDY+FL + G Sbjct: 631 ARQAVENDVHVMGVSSLAAGHKTLIPQVIAELEKLGRPDILVTAGGVIPAQDYDFLYQAG 690 Query: 1446 VSEIFGPGTRIPDAASKVVDAII 1514 V+ IFGPGT + +A+KV++ ++ Sbjct: 691 VAAIFGPGTPVAYSAAKVLEILL 713
>sp|Q05065|MUTB_STRCM Methylmalonyl-CoA mutase large subunit (MCM-alpha) Length = 733 Score = 590 bits (1522), Expect = e-168 Identities = 294/511 (57%), Positives = 376/511 (73%), Gaps = 5/511 (0%) Frame = +3 Query: 3 PSMKIIGDIFEYTAKHMPKFNSISISGYHMQEAGANAILEMAYTIADGLEYCRTGLKAGL 182 PSM+II DIF YT++ MP++NSISISGYH+QEAGA A LE+AYT+ADG+EY R G +AGL Sbjct: 219 PSMRIISDIFAYTSQKMPRYNSISISGYHIQEAGATADLELAYTLADGVEYLRAGQEAGL 278 Query: 183 HIDEFAPRLSFFWGVGMNFYMEIAKMRAARRVWAELLKSKFNSKDKKSLLLRAHSQTSGW 362 +D FAPRLSFFW +GMNF+ME+AK+RAAR +WA+L+K +F+ K+ KSL LR HSQTSGW Sbjct: 279 DVDAFAPRLSFFWAIGMNFFMEVAKLRAARLLWAKLVK-QFDPKNAKSLSLRTHSQTSGW 337 Query: 363 SLTEQDPYNNIMRTTIEAMAAVFGGTQSLHTNAFDEALGLPTKFSARIARNXXXXXXXXX 542 SLT QD +NN+ RT +EAMAA G TQSLHTNA DEAL LPT FSARIARN Sbjct: 338 SLTAQDVFNNVTRTCVEAMAATQGHTQSLHTNALDEALALPTDFSARIARNTQLLIQQES 397 Query: 543 XXXKVADPWAGSFMMESLTQEIYSEAIKVIEEIEAMGGMAKAVSEGIPKLKIEECAAKRQ 722 + DPW GS +E LT ++ A + IEE+EA GGMA+A+ GIPKL++EE AA+ Q Sbjct: 398 GTTRTIDPWGGSAYVEKLTYDLARRAWQHIEEVEAAGGMAQAIDAGIPKLRVEEAAARTQ 457 Query: 723 AHIDSGQEVIVGVNKYRLENPEKVDVLHIDNTKVRESQIAKINALKQSRDNNKAQNILKK 902 A IDSG++ ++GVNKYR++ E++DVL +DN+ VR QI K+ L++ RD+ Q+ L+ Sbjct: 458 ARIDSGRQPVIGVNKYRVDTDEQIDVLKVDNSSVRAQQIEKLRRLREERDDAACQDALRA 517 Query: 903 LEECCRSG-----HGNLLEIAVDASRARCTVGEITDAMVAAYGRHVAVGRLVSGAYKSEF 1067 L G GNLL +AVDA+RA+ TVGEI+DA+ + YGRH R +SG Y++E Sbjct: 518 LTAAAERGPGQGLEGNLLALAVDAARAKATVGEISDALESVYGRHAGQIRTISGVYRTEA 577 Query: 1068 GDNKEIAAVLKKVEDFLKNEGRRPRILIAKLGQDGHDRGGKVIATGFSDLGFDVDVGPLF 1247 G + + V+ F + EGRRPRIL+AK+GQDGHDRG KVIA+ F+DLGFDVDVGPLF Sbjct: 578 GQSPSVERTRALVDAFDEAEGRRPRILVAKMGQDGHDRGQKVIASAFADLGFDVDVGPLF 637 Query: 1248 STPKEAVEQAVDADVHVIGVSSLAAGHMTLVPQLIDELKKAGRSDIVVVAGGVIPPQDYE 1427 TP E QAV+ADVH++GVSSLAAGH+TLVP L +EL GR DI++V GGVIPPQD E Sbjct: 638 QTPAEVARQAVEADVHIVGVSSLAAGHLTLVPALREELAAEGRDDIMIVVGGVIPPQDVE 697 Query: 1428 FLRKVGVSEIFGPGTRIPDAASKVVDAIISN 1520 L + G + +F PGT IPDAA +V + ++ Sbjct: 698 ALHEAGATAVFPPGTVIPDAAHDLVKRLAAD 728
>sp|O86028|MUTB_RHIME Methylmalonyl-CoA mutase (MCM) Length = 712 Score = 567 bits (1462), Expect = e-161 Identities = 285/503 (56%), Positives = 367/503 (72%) Frame = +3 Query: 3 PSMKIIGDIFEYTAKHMPKFNSISISGYHMQEAGANAILEMAYTIADGLEYCRTGLKAGL 182 PSM+I+ DI EYTAK MPKFNSISISGYHMQEAGA + E+A+T+ADG EY R L GL Sbjct: 204 PSMRIVADIIEYTAKEMPKFNSISISGYHMQEAGATLVQELAFTLADGREYVRAALAKGL 263 Query: 183 HIDEFAPRLSFFWGVGMNFYMEIAKMRAARRVWAELLKSKFNSKDKKSLLLRAHSQTSGW 362 ++D+FA RLSFF+ +GMNF+ME AK+RAAR +W +++ +F + SL+LR H QTSG Sbjct: 264 NVDDFAGRLSFFFAIGMNFFMEAAKLRAARLLWTRIMQ-EFKPEKASSLMLRTHCQTSGV 322 Query: 363 SLTEQDPYNNIMRTTIEAMAAVFGGTQSLHTNAFDEALGLPTKFSARIARNXXXXXXXXX 542 SL EQDPYNNI+RT EAM+AV GGTQSLHTN+FDEA+ LPT FSARIARN Sbjct: 323 SLQEQDPYNNIVRTAFEAMSAVLGGTQSLHTNSFDEAMALPTDFSARIARNTQLILQHET 382 Query: 543 XXXKVADPWAGSFMMESLTQEIYSEAIKVIEEIEAMGGMAKAVSEGIPKLKIEECAAKRQ 722 KV DP AGS+ +ESLT E+ +A +IEE+EA+GGM KAV+ G+PK IEE A +RQ Sbjct: 383 GVTKVVDPLAGSYYVESLTNELAEKAWGLIEEVEALGGMTKAVNAGLPKRLIEEAATRRQ 442 Query: 723 AHIDSGQEVIVGVNKYRLENPEKVDVLHIDNTKVRESQIAKINALKQSRDNNKAQNILKK 902 A +D +EVIVGVNKYRLEN + +D+L IDN VR +Q+ +I ++ RD+ K + L Sbjct: 443 AAVDRAEEVIVGVNKYRLENEQPIDILQIDNAAVRTAQVKRIEETRRRRDSQKMKQALDA 502 Query: 903 LEECCRSGHGNLLEIAVDASRARCTVGEITDAMVAAYGRHVAVGRLVSGAYKSEFGDNKE 1082 L + RSG GNLL AV+A+RAR TVGEITDAM A+G + A+ +V+ Y + + E Sbjct: 503 LADVARSGKGNLLAAAVEAARARATVGEITDAMREAFGDYTAIPEVVTDIYGKAYEGDPE 562 Query: 1083 IAAVLKKVEDFLKNEGRRPRILIAKLGQDGHDRGGKVIATGFSDLGFDVDVGPLFSTPKE 1262 + + ++ + K G +P+I++AKLGQDGHDRG KVIA+ F D+GFDV GPLF TP+E Sbjct: 563 LGVLAGRLGEATKRLGHKPKIMVAKLGQDGHDRGAKVIASAFGDIGFDVVAGPLFQTPEE 622 Query: 1263 AVEQAVDADVHVIGVSSLAAGHMTLVPQLIDELKKAGRSDIVVVAGGVIPPQDYEFLRKV 1442 A + A+ +V VIGVSSLAAGH TL+PQL + LKK G DI+VV GGVIP QDY++L + Sbjct: 623 AADLALAEEVTVIGVSSLAAGHRTLMPQLAEALKKRGGEDIIVVCGGVIPRQDYDYLMEN 682 Query: 1443 GVSEIFGPGTRIPDAASKVVDAI 1511 GV+ +FGPGT++ DAA V+D I Sbjct: 683 GVAAVFGPGTQVLDAARAVLDLI 705
>sp|P65487|MUTB_MYCTU Probable methylmalonyl-CoA mutase large subunit (MCM) sp|P65488|MUTB_MYCBO Probable methylmalonyl-CoA mutase large subunit (MCM) Length = 750 Score = 566 bits (1459), Expect = e-161 Identities = 294/504 (58%), Positives = 362/504 (71%), Gaps = 8/504 (1%) Frame = +3 Query: 3 PSMKIIGDIFEYTAKHMPKFNSISISGYHMQEAGANAILEMAYTIADGLEYCRTGLKAGL 182 PSM+II DIF YT+ MPKFNSISISGYH+QEAGA A LE+AYT+ADG++Y R GL AGL Sbjct: 232 PSMRIISDIFAYTSAKMPKFNSISISGYHIQEAGATADLELAYTLADGVDYIRAGLNAGL 291 Query: 183 HIDEFAPRLSFFWGVGMNFYMEIAKMRAARRVWAELLKSKFNSKDKKSLLLRAHSQTSGW 362 ID FAPRLSFFWG+GMNF+ME+AK+RA R +W+EL+ ++F K KSL LR HSQTSGW Sbjct: 292 DIDSFAPRLSFFWGIGMNFFMEVAKLRAGRLLWSELV-AQFAPKSAKSLSLRTHSQTSGW 350 Query: 363 SLTEQDPYNNIMRTTIEAMAAVFGGTQSLHTNAFDEALGLPTKFSARIARNXXXXXXXXX 542 SLT QD +NN+ RT IEAMAA G TQSLHTNA DEAL LPT FSARIARN Sbjct: 351 SLTAQDVFNNVARTCIEAMAATQGHTQSLHTNALDEALALPTDFSARIARNTQLVLQQES 410 Query: 543 XXXKVADPWAGSFMMESLTQEIYSEAIKVIEEIEAMGGMAKAVSEGIPKLKIEECAAKRQ 722 + DPW GS+ +E LT + A I E+ GGMA+A+S+GIPKL+IEE AA+ Q Sbjct: 411 GTTRPIDPWGGSYYVEWLTHRLARRARAHIAEVAEHGGMAQAISDGIPKLRIEEAAARTQ 470 Query: 723 AHIDSGQEVIVGVNKYRLENPEKVDVLHIDNTKVRESQIAKINALKQSRDNNKAQNILKK 902 A IDSGQ+ +VGVNKY++ +++VL ++N++VR Q+AK+ L+ RD + L + Sbjct: 471 ARIDSGQQPVVGVNKYQVPEDHEIEVLKVENSRVRAEQLAKLQRLRAGRDEPAVRAALAE 530 Query: 903 LEECC----RSGH----GNLLEIAVDASRARCTVGEITDAMVAAYGRHVAVGRLVSGAYK 1058 L R+G NLL +A+DA+RA+ TVGEI++A+ YGRH A R +SG Y+ Sbjct: 531 LTRAAAEQGRAGADGLGNNLLALAIDAARAQATVGEISEALEKVYGRHRAEIRTISGVYR 590 Query: 1059 SEFGDNKEIAAVLKKVEDFLKNEGRRPRILIAKLGQDGHDRGGKVIATGFSDLGFDVDVG 1238 E G IAA + VE F + +GRRPRILIAK+GQDGHDRG KVIAT F+D+GFDVDVG Sbjct: 591 DEVGKAPNIAAATELVEKFAEADGRRPRILIAKMGQDGHDRGQKVIATAFADIGFDVDVG 650 Query: 1239 PLFSTPKEAVEQAVDADVHVIGVSSLAAGHMTLVPQLIDELKKAGRSDIVVVAGGVIPPQ 1418 LFSTP+E QA D DVHVIGVSSLAAGH+TLVP L D L + GR DI++V GGVIPP Sbjct: 651 SLFSTPEEVARQAADNDVHVIGVSSLAAGHLTLVPALRDALAQVGRPDIMIVVGGVIPPG 710 Query: 1419 DYEFLRKVGVSEIFGPGTRIPDAA 1490 D++ L G + IF PGT I DAA Sbjct: 711 DFDELYAAGATAIFPPGTVIADAA 734
>sp|P11653|MUTB_PROFR Methylmalonyl-CoA mutase large subunit (MCM-alpha) Length = 728 Score = 566 bits (1459), Expect = e-161 Identities = 286/505 (56%), Positives = 369/505 (73%), Gaps = 5/505 (0%) Frame = +3 Query: 3 PSMKIIGDIFEYTAKHMPKFNSISISGYHMQEAGANAILEMAYTIADGLEYCRTGLKAGL 182 PSM+II +IF YT+ +MPK+NSISISGYHMQEAGA A +EMAYT+ADG++Y R G GL Sbjct: 216 PSMRIISEIFAYTSANMPKWNSISISGYHMQEAGATADIEMAYTLADGVDYIRAGESVGL 275 Query: 183 HIDEFAPRLSFFWGVGMNFYMEIAKMRAARRVWAELLKSKFNSKDKKSLLLRAHSQTSGW 362 ++D+FAPRLSFFWG+GMNF+ME+AK+RAAR +WA+L+ +F K+ KS+ LR HSQTSGW Sbjct: 276 NVDQFAPRLSFFWGIGMNFFMEVAKLRAARMLWAKLVH-QFGPKNPKSMSLRTHSQTSGW 334 Query: 363 SLTEQDPYNNIMRTTIEAMAAVFGGTQSLHTNAFDEALGLPTKFSARIARNXXXXXXXXX 542 SLT QD YNN++RT IEAMAA G TQSLHTN+ DEA+ LPT FSARIARN Sbjct: 335 SLTAQDVYNNVVRTCIEAMAATQGHTQSLHTNSLDEAIALPTDFSARIARNTQLFLQQES 394 Query: 543 XXXKVADPWAGSFMMESLTQEIYSEAIKVIEEIEAMGGMAKAVSEGIPKLKIEECAAKRQ 722 +V DPW+GS +E LT ++ +A I+E+E +GGMAKA+ +GIPK++IEE AA+ Q Sbjct: 395 GTTRVIDPWSGSAYVEELTWDLARKAWGHIQEVEKVGGMAKAIEKGIPKMRIEEAAARTQ 454 Query: 723 AHIDSGQEVIVGVNKYRLENPEKVDVLHIDNTKVRESQIAKINALKQSRDNNKAQNILKK 902 A IDSG++ ++GVNKYRLE+ +DVL +DN+ V Q AK+ L+ RD K + L K Sbjct: 455 ARIDSGRQPLIGVNKYRLEHEPPLDVLKVDNSTVLAEQKAKLVKLRAERDPEKVKAALDK 514 Query: 903 LEECC-----RSGHGNLLEIAVDASRARCTVGEITDAMVAAYGRHVAVGRLVSGAYKSEF 1067 + + NLL++ +DA RA TVGE++DA+ +GR+ A R +SG Y E Sbjct: 515 ITWAAGNPDDKDPDRNLLKLCIDAGRAMATVGEMSDALEKVFGRYTAQIRTISGVYSKEV 574 Query: 1068 GDNKEIAAVLKKVEDFLKNEGRRPRILIAKLGQDGHDRGGKVIATGFSDLGFDVDVGPLF 1247 + E+ + VE+F + EGRRPRIL+AK+GQDGHDRG KVIAT ++DLGFDVDVGPLF Sbjct: 575 KNTPEVEEARELVEEFEQAEGRRPRILLAKMGQDGHDRGQKVIATAYADLGFDVDVGPLF 634 Query: 1248 STPKEAVEQAVDADVHVIGVSSLAAGHMTLVPQLIDELKKAGRSDIVVVAGGVIPPQDYE 1427 TP+E QAV+ADVHV+GVSSLA GH+TLVP L EL K GR DI++ GGVIP QD++ Sbjct: 635 QTPEETARQAVEADVHVVGVSSLAGGHLTLVPALRKELDKLGRPDILITVGGVIPEQDFD 694 Query: 1428 FLRKVGVSEIFGPGTRIPDAASKVV 1502 LRK G EI+ PGT IP++A +V Sbjct: 695 ELRKDGAVEIYTPGTVIPESAISLV 719
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 173,282,547
Number of Sequences: 369166
Number of extensions: 3355378
Number of successful extensions: 9541
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 9029
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 9517
length of database: 68,354,980
effective HSP length: 116
effective length of database: 46,925,720
effective search space used: 20694242520
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail