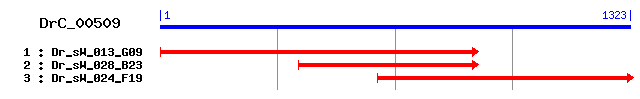
DrC_00509
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00509 (1323 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q91V09|WD13_MOUSE WD-repeat protein 13 299 1e-80 sp|Q6DKP5|WDR13_PANTR WD-repeat protein 13 >gi|20140804|sp|... 295 2e-79 sp|P61964|WDR5_HUMAN WD-repeat protein 5 (BMP2-induced 3-kb... 63 2e-09 sp|P49695|PKWA_THECU Probable serine/threonine-protein kina... 62 5e-09 sp|Q8YRI1|YY46_ANASP Hypothetical WD-repeat protein alr3466 59 4e-08 sp|P39706|SWD1_YEAST COMPASS component SWD1 (Complex protei... 57 9e-08 sp|Q9V3J8|WDS_DROME Will die slowly protein 55 5e-07 sp|Q23256|YH92_CAEEL Hypothetical WD-repeat protein ZC302.2... 55 6e-07 sp|Q15291|RBBP5_HUMAN Retinoblastoma-binding protein 5 (RBB... 52 3e-06 sp|O43017|SWD3_SCHPO Set1 complex component swd3 (Set1C com... 51 7e-06
>sp|Q91V09|WD13_MOUSE WD-repeat protein 13 Length = 485 Score = 299 bits (765), Expect = 1e-80 Identities = 160/353 (45%), Positives = 230/353 (65%), Gaps = 1/353 (0%) Frame = +2 Query: 2 TKIALASRALAGNDSVEEGYAFTGIKHIFD-HCSAGVNRIRFAHNNASRLAIGCEDGSLS 178 T +A ASRA+AG+ S+ E YAF G+ H+FD H V R+RFA+++ RLA DGS+S Sbjct: 139 TSVAEASRAMAGDTSLSENYAFAGMYHVFDQHVDEAVPRVRFANDDRHRLACCSLDGSIS 198 Query: 179 ICCTWPFQEECVLAIMKDQHKSGITDLAWSSANDFILTASRDSTVCLWNVESASLARIYK 358 +C P VL +++ H G++D AWS +ND +++ S D+T+ +W E R Sbjct: 199 LCQLVP-APPTVLHVLRG-HTRGVSDFAWSLSNDILVSTSLDATMRIWASEDGRCIREIP 256 Query: 359 ELNVGKEILTVEFQPMNNNLFVAGGSDGVIQVINLSTGKAVKSGRDKLGGMITALTFEES 538 + + G E+L FQP+NNNL V G + + V+N+STGK VK G KL G + AL+F+ + Sbjct: 257 DPD-GAELLCCTFQPVNNNLTVVGNAKHNVHVMNISTGKKVKGGSSKLTGRVLALSFD-A 314 Query: 539 VGECLWIGNTQGIIQSYLFEVGTGKITRAKRLTVSTKPCTISSLQSRAWINREARDPCLL 718 G LW G+ +G + S+LF++ TGK+T+AKRL V + ++S+ +R+W++REARDP LL Sbjct: 315 PGRLLWAGDDRGSVFSFLFDMATGKLTKAKRLVVH-EGSPVTSISARSWVSREARDPSLL 373 Query: 719 VNASINSLLFFRIASSDGQIVLKNKFPIRHTEKIIHSSFCPLISFRQGACVVTGSEDGNV 898 +NA +N LL +R+ ++G + LK FPI + + S FCPL+SFRQGACVVTGSED V Sbjct: 374 INACLNKLLLYRVVDNEGALQLKRSFPIEQSSHPVRSIFCPLMSFRQGACVVTGSEDMCV 433 Query: 899 LVFDVTAGQLNSPKHCINVLQGHSGAVIDVAVSCDELVLASCDDKGHVILWKR 1057 FDV + K +N LQGHS V+DV+ +CDE +LAS D G VI+W+R Sbjct: 434 HFFDVE----RAAKAAVNKLQGHSAPVLDVSFNCDESLLASSDASGMVIVWRR 482
>sp|Q6DKP5|WDR13_PANTR WD-repeat protein 13 sp|Q9H1Z4|WD13_HUMAN WD-repeat protein 13 Length = 485 Score = 295 bits (756), Expect = 2e-79 Identities = 159/353 (45%), Positives = 228/353 (64%), Gaps = 1/353 (0%) Frame = +2 Query: 2 TKIALASRALAGNDSVEEGYAFTGIKHIFD-HCSAGVNRIRFAHNNASRLAIGCEDGSLS 178 T A ASRA+AG+ S+ E YAF G+ H+FD H V R+RFA+++ RLA DGS+S Sbjct: 139 TSAAEASRAMAGDTSLSENYAFAGMYHVFDQHVDEAVPRVRFANDDRHRLACCSLDGSIS 198 Query: 179 ICCTWPFQEECVLAIMKDQHKSGITDLAWSSANDFILTASRDSTVCLWNVESASLARIYK 358 +C P VL +++ H G++D AWS +ND +++ S D+T+ +W E R Sbjct: 199 LCQLVP-APPTVLRVLRG-HTRGVSDFAWSLSNDILVSTSLDATMRIWASEDGRCIREIP 256 Query: 359 ELNVGKEILTVEFQPMNNNLFVAGGSDGVIQVINLSTGKAVKSGRDKLGGMITALTFEES 538 + + E+L FQP+NNNL V G + + V+N+STGK VK G KL G + AL+F+ + Sbjct: 257 DPD-SAELLCCTFQPVNNNLTVVGNAKHNVHVMNISTGKKVKGGSSKLTGRVLALSFD-A 314 Query: 539 VGECLWIGNTQGIIQSYLFEVGTGKITRAKRLTVSTKPCTISSLQSRAWINREARDPCLL 718 G LW G+ +G + S+LF++ TGK+T+AKRL V + ++S+ +R+W++REARDP LL Sbjct: 315 PGRLLWAGDDRGSVFSFLFDMATGKLTKAKRLVVH-EGSPVTSISARSWVSREARDPSLL 373 Query: 719 VNASINSLLFFRIASSDGQIVLKNKFPIRHTEKIIHSSFCPLISFRQGACVVTGSEDGNV 898 +NA +N LL +R+ ++G + LK FPI + + S FCPL+SFRQGACVVTGSED V Sbjct: 374 INACLNKLLLYRVVDNEGTLQLKRSFPIEQSSHPVRSIFCPLMSFRQGACVVTGSEDMCV 433 Query: 899 LVFDVTAGQLNSPKHCINVLQGHSGAVIDVAVSCDELVLASCDDKGHVILWKR 1057 FDV + K +N LQGHS V+DV+ +CDE +LAS D G VI+W+R Sbjct: 434 HFFDVE----RAAKAAVNKLQGHSAPVLDVSFNCDESLLASSDASGMVIVWRR 482
>sp|P61964|WDR5_HUMAN WD-repeat protein 5 (BMP2-induced 3-kb gene protein) sp|P61965|WDR5_MOUSE WD-repeat protein 5 (BMP2-induced 3-kb gene protein) (WD-repeat protein BIG-3) Length = 334 Score = 62.8 bits (151), Expect = 2e-09 Identities = 65/275 (23%), Positives = 123/275 (44%), Gaps = 2/275 (0%) Frame = +2 Query: 236 HKSGITDLAWSSANDFILTASRDSTVCLWNVESASLARIYKELNVGKEILTVEFQPMNNN 415 HK GI+D+AWSS ++ +++AS D T+ +W+V S + K + F P +N Sbjct: 86 HKLGISDVAWSSDSNLLVSASDDKTLKIWDVSSGKCLKTLK--GHSNYVFCCNFNP-QSN 142 Query: 416 LFVAGGSDGVIQVINLSTGKAVKSGRDKLGGMITALTFEESVGECLWIGNTQGIIQSYLF 595 L V+G D +++ ++ TGK +K+ ++A+ F G + + G+ + ++ Sbjct: 143 LIVSGSFDESVRIWDVKTGKCLKT-LPAHSDPVSAVHFNRD-GSLIVSSSYDGLCR--IW 198 Query: 596 EVGTGKITRAKRLTVSTKPCTISSLQSRAWINREARDPCLLVNASINSLLFFRIASSDGQ 775 + +G+ K L P +++ +L N+L + + Sbjct: 199 DTASGQC--LKTLIDDDNPPV-------SFVKFSPNGKYILAATLDNTLKLWDYSKGK-- 247 Query: 776 IVLKNKFPIRHTEKIIHSSFCPLISFRQGACVVTGSEDGNVLVFDVTAGQLNSPKHCINV 955 LK ++ + I ++F S G +V+GSED V ++++ K + Sbjct: 248 -CLKTYTGHKNEKYCIFANF----SVTGGKWIVSGSEDNLVYIWNL------QTKEIVQK 296 Query: 956 LQGHSGAVIDVAVSCDELVLASC--DDKGHVILWK 1054 LQGH+ VI A E ++AS ++ + LWK Sbjct: 297 LQGHTDVVISTACHPTENIIASAALENDKTIKLWK 331
Score = 39.3 bits (90), Expect = 0.026
Identities = 26/114 (22%), Positives = 45/114 (39%), Gaps = 7/114 (6%)
Frame = +2
Query: 164 DGSL-------SICCTWPFQEECVLAIMKDQHKSGITDLAWSSANDFILTASRDSTVCLW 322
DGSL +C W L + D ++ + +S +IL A+ D+T+ LW
Sbjct: 182 DGSLIVSSSYDGLCRIWDTASGQCLKTLIDDDNPPVSFVKFSPNGKYILAATLDNTLKLW 241
Query: 323 NVESASLARIYKELNVGKEILTVEFQPMNNNLFVAGGSDGVIQVINLSTGKAVK 484
+ + Y K + F V+G D ++ + NL T + V+
Sbjct: 242 DYSKGKCLKTYTGHKNEKYCIFANFSVTGGKWIVSGSEDNLVYIWNLQTKEIVQ 295
>sp|P49695|PKWA_THECU Probable serine/threonine-protein kinase pkwA Length = 742 Score = 61.6 bits (148), Expect = 5e-09 Identities = 66/274 (24%), Positives = 125/274 (45%), Gaps = 2/274 (0%) Frame = +2 Query: 236 HKSGITDLAWSSANDFILTASRDSTVCLWNVESASLARIYKELNVGKEILTVEFQPMNNN 415 H + +A+S + + S D+TV LW+V +A +++ +L + F P + + Sbjct: 500 HTDWVRAVAFSPDGALLASGSDDATVRLWDVAAAEERAVFEGHT--HYVLDIAFSP-DGS 556 Query: 416 LFVAGGSDGVIQVINLSTGK--AVKSGRDKLGGMITALTFEESVGECLWIGNTQGIIQSY 589 + +G DG ++ N++TG AV G + A+ F G + G+ G I+ Sbjct: 557 MVASGSRDGTARLWNVATGTEHAVLKGHTDY---VYAVAFSPD-GSMVASGSRDGTIR-- 610 Query: 590 LFEVGTGKITRAKRLTVSTKPCTISSLQSRAWINREARDPCLLVNASINSLLFFRIASSD 769 L++V TGK +R + + SL + D +LV+ S +++ + +AS + Sbjct: 611 LWDVATGK----ERDVLQAPAENVVSLAF-------SPDGSMLVHGSDSTVHLWDVASGE 659 Query: 770 GQIVLKNKFPIRHTEKIIHSSFCPLISFRQGACVVTGSEDGNVLVFDVTAGQLNSPKHCI 949 + HT+ + +F P GA + +GS+D + ++DV A + ++ Sbjct: 660 ALHTFEG-----HTDWVRAVAFSP-----DGALLASGSDDRTIRLWDVAAQEEHT----- 704 Query: 950 NVLQGHSGAVIDVAVSCDELVLASCDDKGHVILW 1051 L+GH+ V VA + LAS + G + +W Sbjct: 705 -TLEGHTEPVHSVAFHPEGTTLASASEDGTIRIW 737
Score = 48.5 bits (114), Expect = 4e-05
Identities = 62/285 (21%), Positives = 122/285 (42%), Gaps = 7/285 (2%)
Frame = +2
Query: 80 HIFDHCSAGVNRIRFAHNNASRLAIGCEDGSLSICCTWPFQEECVLAIMKDQHKSGITDL 259
H + + V + F+ + A LA G +D ++ + +E V + H + D+
Sbjct: 495 HTLEGHTDWVRAVAFSPDGAL-LASGSDDATVRLWDVAAAEERAVF----EGHTHYVLDI 549
Query: 260 AWSSANDFILTASRDSTVCLWNVESASLARIYKELNVGKEILTVEFQPMNNNLFVAGGSD 439
A+S + + SRD T LWNV + + + K + V F P + ++ +G D
Sbjct: 550 AFSPDGSMVASGSRDGTARLWNVATGTEHAVLK--GHTDYVYAVAFSP-DGSMVASGSRD 606
Query: 440 GVIQVINLSTGKAVKSGRDKL---GGMITALTFEESVGECLWIGNTQGIIQSYLFEVGTG 610
G I++ +++TGK RD L + +L F G L G+ + +L++V +G
Sbjct: 607 GTIRLWDVATGKE----RDVLQAPAENVVSLAFSPD-GSMLVHGSDSTV---HLWDVASG 658
Query: 611 KITRAKRLTVSTKPCTISSLQSRA-WINREARDP--CLLVNASIN-SLLFFRIASSDGQI 778
+ + + + W+ A P LL + S + ++ + +A+ +
Sbjct: 659 E--------------ALHTFEGHTDWVRAVAFSPDGALLASGSDDRTIRLWDVAAQEEHT 704
Query: 779 VLKNKFPIRHTEKIIHSSFCPLISFRQGACVVTGSEDGNVLVFDV 913
L+ HTE + +F P +G + + SEDG + ++ +
Sbjct: 705 TLEG-----HTEPVHSVAFHP-----EGTTLASASEDGTIRIWPI 739
Score = 33.5 bits (75), Expect = 1.4
Identities = 23/64 (35%), Positives = 33/64 (51%)
Frame = +2
Query: 860 GACVVTGSEDGNVLVFDVTAGQLNSPKHCINVLQGHSGAVIDVAVSCDELVLASCDDKGH 1039
G+ + GS D + V+DV +G ++ L+GH+ V VA S D +LAS D
Sbjct: 471 GSLLAGGSGDKLIHVWDVASGDE------LHTLEGHTDWVRAVAFSPDGALLASGSDDAT 524
Query: 1040 VILW 1051
V LW
Sbjct: 525 VRLW 528
>sp|Q8YRI1|YY46_ANASP Hypothetical WD-repeat protein alr3466 Length = 1526 Score = 58.5 bits (140), Expect = 4e-08 Identities = 78/341 (22%), Positives = 145/341 (42%), Gaps = 26/341 (7%) Frame = +2 Query: 107 VNRIRFAHNNASRLAIGCEDGSLSICCTWPFQEECVLAIMKDQHKSGITDLAWSSANDFI 286 VN + F+ + A+ LA G D ++ + W L I++ H S + + ++ + Sbjct: 1161 VNAVAFSPDGAT-LASGSGDQTVRL---WDISSSKCLYILQG-HTSWVNSVVFNPDGSTL 1215 Query: 287 LTASRDSTVCLWNVESASLARIYKELNVGKEILTVEFQPMNNNLFVAGGSDGVIQVINLS 466 + S D TV LW + S+ ++ + +V F P + ++ +G SD +++ ++S Sbjct: 1216 ASGSSDQTVRLWEINSSKCLCTFQGHT--SWVNSVVFNP-DGSMLASGSSDKTVRLWDIS 1272 Query: 467 TGKAVKSGRDKL-----------GGMI-------TALTFEESVGECL--WIGNTQGIIQS 586 + K + + + G M+ T +E S +CL + G+T + Sbjct: 1273 SSKCLHTFQGHTNWVNSVAFNPDGSMLASGSGDQTVRLWEISSSKCLHTFQGHTSWVSSV 1332 Query: 587 YLFEVGT----GKITRAKRL-TVSTKPCTISSLQSRAWINREARDPCLLVNASINSLLFF 751 GT G + RL ++S+ C + L W+ P + AS + Sbjct: 1333 TFSPDGTMLASGSDDQTVRLWSISSGECLYTFLGHTNWVGSVIFSPDGAILASGSGDQTV 1392 Query: 752 RIAS-SDGQIVLKNKFPIRHTEKIIHSSFCPLISFRQGACVVTGSEDGNVLVFDVTAGQL 928 R+ S S G+ + + H + F P G + +GS+D V ++++++G+ Sbjct: 1393 RLWSISSGKCLYTLQ---GHNNWVGSIVFSP-----DGTLLASGSDDQTVRLWNISSGE- 1443 Query: 929 NSPKHCINVLQGHSGAVIDVAVSCDELVLASCDDKGHVILW 1051 C+ L GH +V VA S D L+LAS D + LW Sbjct: 1444 -----CLYTLHGHINSVRSVAFSSDGLILASGSDDETIKLW 1479
Score = 50.8 bits (120), Expect = 9e-06
Identities = 59/272 (21%), Positives = 103/272 (37%)
Frame = +2
Query: 236 HKSGITDLAWSSANDFILTASRDSTVCLWNVESASLARIYKELNVGKEILTVEFQPMNNN 415
H S + + +S + + S D TV LW++ S + +K + +V F P N+
Sbjct: 905 HNSWVNSVGFSQDGKMLASGSDDQTVRLWDISSGQCLKTFK--GHTSRVRSVVFSP-NSL 961
Query: 416 LFVAGGSDGVIQVINLSTGKAVKSGRDKLGGMITALTFEESVGECLWIGNTQGIIQSYLF 595
+ +G SD +++ ++S+G+ + F+ G W+ + + +
Sbjct: 962 MLASGSSDQTVRLWDISSGECL-------------YIFQGHTG---WVYSVAFNLDGSML 1005
Query: 596 EVGTGKITRAKRLTVSTKPCTISSLQSRAWINREARDPCLLVNASINSLLFFRIASSDGQ 775
G+G T + ISS Q C + S + + SSDG
Sbjct: 1006 ATGSGDQT--------VRLWDISSSQ------------CFYIFQGHTSCVRSVVFSSDG- 1044
Query: 776 IVLKNKFPIRHTEKIIHSSFCPLISFRQGACVVTGSEDGNVLVFDVTAGQLNSPKHCINV 955
A + +GS+D V ++D+++G +C+
Sbjct: 1045 -----------------------------AMLASGSDDQTVRLWDISSG------NCLYT 1069
Query: 956 LQGHSGAVIDVAVSCDELVLASCDDKGHVILW 1051
LQGH+ V V S D +LAS D V LW
Sbjct: 1070 LQGHTSCVRSVVFSPDGAMLASGGDDQIVRLW 1101
Score = 50.8 bits (120), Expect = 9e-06
Identities = 80/346 (23%), Positives = 140/346 (40%), Gaps = 30/346 (8%)
Frame = +2
Query: 107 VNRIRFAHNNASRLAIGCEDGSLSICCTWPFQEECVLAIMKDQHKSGITDLAWSSANDFI 286
VN + F+ + LA G +D ++ + W L K H S + + +S + +
Sbjct: 909 VNSVGFSQDG-KMLASGSDDQTVRL---WDISSGQCLKTFKG-HTSRVRSVVFSPNSLML 963
Query: 287 LTASRDSTVCLWNVESASLARIYKE----------------LNVGKEILTVEFQPMNNN- 415
+ S D TV LW++ S I++ L G TV ++++
Sbjct: 964 ASGSSDQTVRLWDISSGECLYIFQGHTGWVYSVAFNLDGSMLATGSGDQTVRLWDISSSQ 1023
Query: 416 --LFVAGGSDGVIQVINLSTGKAVKSGRDKLGGMITALTFEESVGECLWI--GNTQGIIQ 583
G + V V+ S G + SG D T ++ S G CL+ G+T ++
Sbjct: 1024 CFYIFQGHTSCVRSVVFSSDGAMLASGSDDQ----TVRLWDISSGNCLYTLQGHTS-CVR 1078
Query: 584 SYLFE-----VGTGKITRAKRL-TVSTKPCTISSLQSRAWINREARDP--CLLVNASINS 739
S +F + +G + RL +S+ C + +W+ P L N S +
Sbjct: 1079 SVVFSPDGAMLASGGDDQIVRLWDISSGNCLYTLQGYTSWVRFLVFSPNGVTLANGSSDQ 1138
Query: 740 LL-FFRIASSDGQIVLKNKFPIRHTEKIIHSSFCPLISFRQGACVVTGSEDGNVLVFDVT 916
++ + I+S L+ HT + +F P GA + +GS D V ++D++
Sbjct: 1139 IVRLWDISSKKCLYTLQG-----HTNWVNAVAFSP-----DGATLASGSGDQTVRLWDIS 1188
Query: 917 AGQLNSPKHCINVLQGHSGAVIDVAVSCDELVLASCDDKGHVILWK 1054
+ + C+ +LQGH+ V V + D LAS V LW+
Sbjct: 1189 SSK------CLYILQGHTSWVNSVVFNPDGSTLASGSSDQTVRLWE 1228
Score = 50.8 bits (120), Expect = 9e-06
Identities = 77/363 (21%), Positives = 149/363 (41%), Gaps = 38/363 (10%)
Frame = +2
Query: 80 HIFDHCSAGVNRIRFAHNNASRLAIGCEDGSLSICCTWPFQEECVLAIMKDQHKSGITDL 259
+IF + V + F + + S LA G D ++ + W I + H S + +
Sbjct: 984 YIFQGHTGWVYSVAF-NLDGSMLATGSGDQTVRL---WDISSSQCFYIFQG-HTSCVRSV 1038
Query: 260 AWSSANDFILTASRDSTVCLWNVESASLARIYKELNVGKEILTVEFQPMNNNLFVAGGSD 439
+SS + + S D TV LW++ S + +Y + +V F P + + +GG D
Sbjct: 1039 VFSSDGAMLASGSDDQTVRLWDISSGNC--LYTLQGHTSCVRSVVFSP-DGAMLASGGDD 1095
Query: 440 GVIQVINLSTGKAVKSGRDKLGGMITALTFEESVGECLWIGNTQGIIQSYLFEVGTGKIT 619
++++ ++S+G + + + + L F + G L G++ I++ +
Sbjct: 1096 QIVRLWDISSGNCLYTLQGYTS-WVRFLVFSPN-GVTLANGSSDQIVRLW---------- 1143
Query: 620 RAKRLTVSTKPCTISSLQSRAWINREARDP-----------------------CLLV--- 721
+S+K C + W+N A P CL +
Sbjct: 1144 -----DISSKKCLYTLQGHTNWVNAVAFSPDGATLASGSGDQTVRLWDISSSKCLYILQG 1198
Query: 722 -NASINSLLF------FRIASSDGQIVLKNKFPIRHTEKII----HSSFCPLISFR-QGA 865
+ +NS++F SSD + L + I ++ + H+S+ + F G+
Sbjct: 1199 HTSWVNSVVFNPDGSTLASGSSDQTVRL---WEINSSKCLCTFQGHTSWVNSVVFNPDGS 1255
Query: 866 CVVTGSEDGNVLVFDVTAGQLNSPKHCINVLQGHSGAVIDVAVSCDELVLASCDDKGHVI 1045
+ +GS D V ++D+++ + C++ QGH+ V VA + D +LAS V
Sbjct: 1256 MLASGSSDKTVRLWDISSSK------CLHTFQGHTNWVNSVAFNPDGSMLASGSGDQTVR 1309
Query: 1046 LWK 1054
LW+
Sbjct: 1310 LWE 1312
>sp|P39706|SWD1_YEAST COMPASS component SWD1 (Complex proteins associated with SET1 protein SWD1) (Set1C component SWD1) Length = 426 Score = 57.4 bits (137), Expect = 9e-08 Identities = 73/324 (22%), Positives = 131/324 (40%), Gaps = 19/324 (5%) Frame = +2 Query: 146 LAIGCEDGSLSICCTWPFQEECVLAIMKDQHKSGITDLAWSSANDFILTASRDSTVCLWN 325 LA+GC +G+L I F+ CV M H IT +AWS +LT+SRD ++ LW+ Sbjct: 41 LALGCANGALVIYDMDTFRPICVPGNMLGAHVRPITSIAWSPDGRLLLTSSRDWSIKLWD 100 Query: 326 VESASLARIYKELNVGKEILTVEFQPMNNNLFVAG------------GSDGVIQVINLST 469 + S + KE+ I ++ L VA +D V +++ S Sbjct: 101 LSKPS--KPLKEIRFDSPIWGCQWLDAKRRLCVATIFEESDAYVIDFSNDPVASLLSKSD 158 Query: 470 GKAVKSGRDKLGGMITALTFEESVGECLWIGNTQGIIQSYLFE--VGTGKITRAKRLTVS 643 K + S D G + T + +G ++G + Y F T I K + + Sbjct: 159 EKQLSSTPDH--GYVLVCTVHTKHPNIIIVGTSKGWLDFYKFHSLYQTECIHSLKITSSN 216 Query: 644 TKPCTISSLQSRAWINREARDPCLLVNASINSLLFFRIASSDGQIVLKNKFPIRHTEKII 823 K +S R L +N S ++ + I+ D ++ ++ + I Sbjct: 217 IKHLIVSQNGER-----------LAINCSDRTIRQYEISIDDENSAVELTLEHKYQDVIN 265 Query: 824 HSSF-CPLISFRQGACVVT---GSEDGNVLVFDVTAGQLNSPKHCINVLQGHSGAVIDVA 991 + C L S +V GS + +++ T+G L + VL+G +ID+ Sbjct: 266 KLQWNCILFSNNTAEYLVASTHGSSAHELYIWETTSGTL------VRVLEGAEEELIDIN 319 Query: 992 VSCDEL-VLASCDDKGHVILWKRV 1060 + ++++ + G+V +W V Sbjct: 320 WDFYSMSIVSNGFESGNVYVWSVV 343
>sp|Q9V3J8|WDS_DROME Will die slowly protein Length = 361 Score = 55.1 bits (131), Expect = 5e-07 Identities = 61/275 (22%), Positives = 121/275 (44%), Gaps = 2/275 (0%) Frame = +2 Query: 236 HKSGITDLAWSSANDFILTASRDSTVCLWNVESASLARIYKELNVGKEILTVEFQPMNNN 415 HK GI+D+AWSS + +++ S D T+ +W + + + K + F P +N Sbjct: 113 HKLGISDVAWSSDSRLLVSGSDDKTLKVWELSTGKSLKTLK--GHSNYVFCCNFNP-QSN 169 Query: 416 LFVAGGSDGVIQVINLSTGKAVKSGRDKLGGMITALTFEESVGECLWIGNTQGIIQSYLF 595 L V+G D +++ ++ TGK +K+ ++A+ F G + + G+ + ++ Sbjct: 170 LIVSGSFDESVRIWDVRTGKCLKT-LPAHSDPVSAVHFNRD-GSLIVSSSYDGLCR--IW 225 Query: 596 EVGTGKITRAKRLTVSTKPCTISSLQSRAWINREARDPCLLVNASINSLLFFRIASSDGQ 775 + +G+ K L P +++ +L N+L + + Sbjct: 226 DTASGQC--LKTLIDDDNPPV-------SFVKFSPNGKYILAATLDNTLKLWDYSKGK-- 274 Query: 776 IVLKNKFPIRHTEKIIHSSFCPLISFRQGACVVTGSEDGNVLVFDVTAGQLNSPKHCINV 955 LK ++ + I ++F S G +V+GSED V ++++ + K + Sbjct: 275 -CLKTYTGHKNEKYCIFANF----SVTGGKWIVSGSEDNMVYIWNLQS------KEVVQK 323 Query: 956 LQGHSGAVIDVAVSCDELVLASC--DDKGHVILWK 1054 LQGH+ V+ A E ++AS ++ + LWK Sbjct: 324 LQGHTDTVLCTACHPTENIIASAALENDKTIKLWK 358
Score = 38.1 bits (87), Expect = 0.057
Identities = 25/114 (21%), Positives = 45/114 (39%), Gaps = 7/114 (6%)
Frame = +2
Query: 164 DGSL-------SICCTWPFQEECVLAIMKDQHKSGITDLAWSSANDFILTASRDSTVCLW 322
DGSL +C W L + D ++ + +S +IL A+ D+T+ LW
Sbjct: 209 DGSLIVSSSYDGLCRIWDTASGQCLKTLIDDDNPPVSFVKFSPNGKYILAATLDNTLKLW 268
Query: 323 NVESASLARIYKELNVGKEILTVEFQPMNNNLFVAGGSDGVIQVINLSTGKAVK 484
+ + Y K + F V+G D ++ + NL + + V+
Sbjct: 269 DYSKGKCLKTYTGHKNEKYCIFANFSVTGGKWIVSGSEDNMVYIWNLQSKEVVQ 322
>sp|Q23256|YH92_CAEEL Hypothetical WD-repeat protein ZC302.2 in chromosome V Length = 501 Score = 54.7 bits (130), Expect = 6e-07 Identities = 75/339 (22%), Positives = 143/339 (42%), Gaps = 4/339 (1%) Frame = +2 Query: 53 EGYAFTGIKHIFDHCSAGVNRIRFAHNNASRLAIGCEDGSLSICCTWPFQEECVLAIMKD 232 E F+ +K I H + V+ I+F++ L G D + + T LA Sbjct: 199 ENGEFSLVKTISGHTKS-VSVIKFSYCG-KYLGTGSADKQIKVWNTVDMTYLQTLA---- 252 Query: 233 QHKSGITDLAWSSANDFILTASRDSTVCLWNVESASLARIYKELNVGKEILTVEFQPMNN 412 H+ GI D +WSS + FI +AS D+TV +++V S + R + + F P + Sbjct: 253 SHQLGINDFSWSSNSQFIASASDDTTVKIFDVISGACLRTMR--GHTNYVFCCSFNP-QS 309 Query: 413 NLFVAGGSDGVIQVINLSTGKAVKSGRDKLGGMITALTFEESVGECLWIGNTQGIIQSYL 592 +L + G D ++V + TG VK IT++++ G + + G I+ + Sbjct: 310 SLIASAGFDETVRVWDFKTGLCVKC-IPAHSDPITSISYNHD-GNTMATSSYDGCIR--V 365 Query: 593 FEVGTGKITRAKRLTVSTKPCTISSLQSRAWINREARDPCLLVNASINSLLFFRIASSDG 772 ++ +G + V T ++ + C N LL ++ SS Sbjct: 366 WDAASGSCLKT---LVDTDHAPVTFV-------------CFSPNGKY--LLSAQLDSSLK 407 Query: 773 QIVLKNKFPIRHTEKIIHSSFCPL--ISFRQGACVVTGSEDGNVLVFDVTAGQLNSPKHC 946 K P+++ + +C +S G +++GSEDG +LV+ + Q+ Sbjct: 408 LWDPKKAKPLKYYNGHKNKKYCLFANMSVPLGKHIISGSEDGRILVWSIQTKQI------ 461 Query: 947 INVLQGHSGAVI--DVAVSCDELVLASCDDKGHVILWKR 1057 + +L+GH+ V+ D + + + + + +W+R Sbjct: 462 VQILEGHTTPVLATDSHPTLNIIASGGLEPDNVIRIWRR 500
>sp|Q15291|RBBP5_HUMAN Retinoblastoma-binding protein 5 (RBBP-5) (Retinoblastoma-binding protein RBQ-3) Length = 538 Score = 52.4 bits (124), Expect = 3e-06 Identities = 70/310 (22%), Positives = 131/310 (42%), Gaps = 8/310 (2%) Frame = +2 Query: 146 LAIGCEDGSLSICCTWPFQEECVLAIMKDQHKSGITDLAWSSANDFILTASRDSTVCLWN 325 LA+GC DG + I W F + I+ H + L WS +++AS D+ V W+ Sbjct: 39 LAVGCNDGRIVI---WDFLTRGIAKIIS-AHIHPVCSLCWSRDGHKLVSASTDNIVSQWD 94 Query: 326 VESASLARIYKELNVGKEILTVEFQPMNNNLFVAGGSDGVIQVINLSTGKAVKSGRDKLG 505 V S + ++ IL V++ P + N + ++ LS K V D Sbjct: 95 VLSGDCDQRFR---FPSPILKVQYHPRDQNKVLVCPMKSAPVMLTLSDSKHVVLPVDDDS 151 Query: 506 GMITALTFEESVGECLWIGNTQGIIQSYLFEVGTGKITRAKRLTVSTKPCTISSLQSRAW 685 + +F+ GE ++ GN +G I + + + + + R+T T T +++S Sbjct: 152 DLNVVASFDRR-GEYIYTGNAKGKI--LVLKTDSQDLVASFRVTTGTSNTT--AIKS--- 203 Query: 686 INREARDPCLLVNASINSLLFFRIASSDGQIVLK-----NKFPIRHTEKIIHSSFCPLIS 850 I + C L+N + + + DG+ +L P++ + +++ + Sbjct: 204 IEFARKGSCFLINTADRIIRVY-----DGREILTCGRDGEPEPMQELQDLVNRTPWKKCC 258 Query: 851 FR-QGACVVTGSEDGNVL-VFDVTAGQLNSPKHCINVLQGHSG-AVIDVAVSCDELVLAS 1021 F G +V GS + L +++ + G L + +L G G ++DVA ++AS Sbjct: 259 FSGDGEYIVAGSARQHALYIWEKSIGNL------VKILHGTRGELLLDVAWHPVRPIIAS 312 Query: 1022 CDDKGHVILW 1051 G V +W Sbjct: 313 I-SSGVVSIW 321
>sp|O43017|SWD3_SCHPO Set1 complex component swd3 (Set1C component swd3) (COMPASS component swd3) (Complex proteins associated with set1 protein swd3) Length = 380 Score = 51.2 bits (121), Expect = 7e-06 Identities = 30/117 (25%), Positives = 56/117 (47%) Frame = +2 Query: 134 NASRLAIGCEDGSLSICCTWPFQEECVLAIMKDQHKSGITDLAWSSANDFILTASRDSTV 313 N +A DG++ I F+ EC L H GI+ + W++ + ++ +AS D T+ Sbjct: 65 NKRWIATSSSDGTIKIWSALTFRLECTLF----GHYRGISQVKWATGSKYLASASDDKTI 120 Query: 314 CLWNVESASLARIYKELNVGKEILTVEFQPMNNNLFVAGGSDGVIQVINLSTGKAVK 484 +W+ E R K + +++F P+ L V+G D +++ NL G ++ Sbjct: 121 RIWDFEKRCSVRCLK--GHTNYVSSIDFNPL-GTLLVSGSWDETVRIWNLQDGTCLR 174
Score = 36.6 bits (83), Expect = 0.17
Identities = 23/82 (28%), Positives = 38/82 (46%)
Frame = +2
Query: 806 HTEKIIHSSFCPLISFRQGACVVTGSEDGNVLVFDVTAGQLNSPKHCINVLQGHSGAVID 985
HT + F PL G +V+GS D V ++++ G C+ +L HS +I
Sbjct: 137 HTNYVSSIDFNPL-----GTLLVSGSWDETVRIWNLQDGT------CLRMLPAHSEPIIS 185
Query: 986 VAVSCDELVLASCDDKGHVILW 1051
V++S D + A+ G +W
Sbjct: 186 VSISADGTLCATASYDGMARIW 207
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 145,291,058
Number of Sequences: 369166
Number of extensions: 2998237
Number of successful extensions: 8682
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 7104
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 8502
length of database: 68,354,980
effective HSP length: 114
effective length of database: 47,295,190
effective search space used: 15418231940
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail