DrC_00432
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
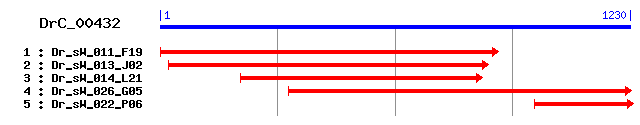
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00432 (1223 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q9GP32|ALF_ECHMU Fructose-bisphosphate aldolase 535 e-151 sp|P53442|ALF_SCHMA Fructose-bisphosphate aldolase 514 e-145 sp|P07764|ALF_DROME Fructose-bisphosphate aldolase 464 e-130 sp|P05063|ALDOC_MOUSE Fructose-bisphosphate aldolase C (Bra... 462 e-130 sp|Q9GKW3|ALDOC_MACFA Fructose-bisphosphate aldolase C (Bra... 461 e-129 sp|P09972|ALDOC_HUMAN Fructose-bisphosphate aldolase C (Bra... 459 e-129 sp|Q5R1X4|ALDOC_PANTR Fructose-bisphosphate aldolase C (Bra... 457 e-128 sp|P09117|ALDOC_RAT Fructose-bisphosphate aldolase C (Brain... 456 e-128 sp|P53445|ALF1_LAMJA Fructose-bisphosphate aldolase, muscle... 455 e-127 sp|P05065|ALDOA_RAT Fructose-bisphosphate aldolase A (Muscl... 448 e-125
>sp|Q9GP32|ALF_ECHMU Fructose-bisphosphate aldolase Length = 363 Score = 535 bits (1378), Expect = e-151 Identities = 261/363 (71%), Positives = 302/363 (83%) Frame = +3 Query: 15 MPEFARYLSEHQLKELKDIANAIVSPGKGILAADESTGTLGKRFKGIHLDNTETNRRQYR 194 M F YL ++KEL++ A+AIV+PGKG+LAADEST T+GKRF I+L+NTE NRR YR Sbjct: 1 MSRFVPYLCAEKMKELRENASAIVAPGKGLLAADESTNTIGKRFAAINLENTEENRRAYR 60 Query: 195 QLLFATDPILARNISGVILYHETLHQKGDDGQPLVKYLQDRGIIPGIKVDTGCVPLLGTF 374 +LLF TDP A++ISGVIL+HET++QK DG+P V+ L++RG++PGIKVD G VPL GT Sbjct: 61 ELLFTTDPEFAKHISGVILFHETVYQKTKDGKPFVELLRERGVLPGIKVDLGVVPLGGTA 120 Query: 375 DECTTQGLDGLDERCAIYKKDGCHFAKWRCVLKIGQHTPSYLAMIENANVLARYAMICQK 554 DECTTQGLD L +RCA Y DGC FAKWRCVLKI H PSYLAM+ENANVLARYA ICQ+ Sbjct: 121 DECTTQGLDNLAQRCAQYYNDGCRFAKWRCVLKISSHNPSYLAMLENANVLARYAFICQQ 180 Query: 555 NGLVPIVEPEVLPDGTHDLFTAQRVTEDVLAFVYKALADHHIYLEGTLLKPNMVTAGMQS 734 NGLVPIVEPEVLPDG HDL TAQRVTE VL+FVYKALADHH+YLEGTLLKPNMVT G Sbjct: 181 NGLVPIVEPEVLPDGDHDLETAQRVTEQVLSFVYKALADHHVYLEGTLLKPNMVTCGQSC 240 Query: 735 PRKYCPEQNAQATVEALMRRVPPAVPGITFLSGGMSELEATLNLNAINRYPGKKPWALTF 914 +KY E NA+ATVEAL R VP AVPG+ FLSGG SEL+AT NLNAIN+YPGKKPWAL+F Sbjct: 241 TKKYSVEDNARATVEALQRTVPVAVPGVVFLSGGQSELDATRNLNAINKYPGKKPWALSF 300 Query: 915 SFGRALQASVIQRWKGKAENIIPAQMELLKLAQANGAASLGRFEGQLETAAGGQSLFVAN 1094 SFGRALQAS I W+GK EN+ Q E L+LA+ANGAASLG+FEG+L+TAAG +SLFVAN Sbjct: 301 SFGRALQASAIAAWQGKPENVKAGQAEFLQLAKANGAASLGKFEGELKTAAGQKSLFVAN 360 Query: 1095 HAY 1103 HAY Sbjct: 361 HAY 363
>sp|P53442|ALF_SCHMA Fructose-bisphosphate aldolase Length = 363 Score = 514 bits (1323), Expect = e-145 Identities = 260/363 (71%), Positives = 290/363 (79%) Frame = +3 Query: 15 MPEFARYLSEHQLKELKDIANAIVSPGKGILAADESTGTLGKRFKGIHLDNTETNRRQYR 194 M F YL+E Q +L+ IA AI +PGKGILAADEST T+GKR + I ++N E NRR YR Sbjct: 1 MSRFQPYLTEAQENDLRRIAQAICAPGKGILAADESTATMGKRLQQIGVENNEENRRLYR 60 Query: 195 QLLFATDPILARNISGVILYHETLHQKGDDGQPLVKYLQDRGIIPGIKVDTGCVPLLGTF 374 QLLF+ D LA NISGVIL+ ETLHQK DDG+ L L +R IIPGIKVD G VPL GT Sbjct: 61 QLLFSADHKLAENISGVILFEETLHQKSDDGKTLPTLLAERNIIPGIKVDKGVVPLAGTD 120 Query: 375 DECTTQGLDGLDERCAIYKKDGCHFAKWRCVLKIGQHTPSYLAMIENANVLARYAMICQK 554 +E TTQGLD L RCA Y + GC FAKWRCVLKI HTPSYLAM+ENANVLARYA ICQ+ Sbjct: 121 NETTTQGLDDLASRCAEYWRLGCRFAKWRCVLKISSHTPSYLAMLENANVLARYASICQQ 180 Query: 555 NGLVPIVEPEVLPDGTHDLFTAQRVTEDVLAFVYKALADHHIYLEGTLLKPNMVTAGMQS 734 NGLVPIVEPEVLPDG HDL TAQRVTE VLAFVYKALADHH+YLEGTLLKPNMVTAG Sbjct: 181 NGLVPIVEPEVLPDGDHDLLTAQRVTEQVLAFVYKALADHHVYLEGTLLKPNMVTAGQAC 240 Query: 735 PRKYCPEQNAQATVEALMRRVPPAVPGITFLSGGMSELEATLNLNAINRYPGKKPWALTF 914 + Y P++NA ATV AL R VPPAVPGITFLSGG SEL+AT NLN IN+ PG KPWALTF Sbjct: 241 KKAYTPQENALATVRALQRTVPPAVPGITFLSGGQSELDATKNLNEINKIPGPKPWALTF 300 Query: 915 SFGRALQASVIQRWKGKAENIIPAQMELLKLAQANGAASLGRFEGQLETAAGGQSLFVAN 1094 SFGRALQASV+ WKGK EN+ AQ ELLKLA+ANGAA++G+FEG + T G +SLFVAN Sbjct: 301 SFGRALQASVLATWKGKKENVHAAQEELLKLAKANGAAAVGKFEGNMGTTLGDKSLFVAN 360 Query: 1095 HAY 1103 HAY Sbjct: 361 HAY 363
>sp|P07764|ALF_DROME Fructose-bisphosphate aldolase Length = 361 Score = 464 bits (1194), Expect = e-130 Identities = 242/363 (66%), Positives = 281/363 (77%) Frame = +3 Query: 15 MPEFARYLSEHQLKELKDIANAIVSPGKGILAADESTGTLGKRFKGIHLDNTETNRRQYR 194 M + Y S+ EL++IA IV+PGKGILAADES T+GKR + I ++NTE NRR YR Sbjct: 1 MTTYFNYPSKELQDELREIAQKIVAPGKGILAADESGPTMGKRLQDIGVENTEDNRRAYR 60 Query: 195 QLLFATDPILARNISGVILYHETLHQKGDDGQPLVKYLQDRGIIPGIKVDTGCVPLLGTF 374 QLLF+TDP LA NISGVIL+HETL+QK DDG P + L+ +GII GIKVD G VPL G+ Sbjct: 61 QLLFSTDPKLAENISGVILFHETLYQKADDGTPFAEILKKKGIILGIKVDKGVVPLFGSE 120 Query: 375 DECTTQGLDGLDERCAIYKKDGCHFAKWRCVLKIGQHTPSYLAMIENANVLARYAMICQK 554 DE TTQGLD L RCA YKKDGC FAKWRCVLKIG++TPSY +++ENANVLARYA ICQ Sbjct: 121 DEVTTQGLDDLAARCAQYKKDGCDFAKWRCVLKIGKNTPSYQSILENANVLARYASICQS 180 Query: 555 NGLVPIVEPEVLPDGTHDLFTAQRVTEDVLAFVYKALADHHIYLEGTLLKPNMVTAGMQS 734 +VPIVEPEVLPDG HDL AQ+VTE VLA VYKAL+DHH+YLEGTLLKPNMVTAG QS Sbjct: 181 QRIVPIVEPEVLPDGDHDLDRAQKVTETVLAAVYKALSDHHVYLEGTLLKPNMVTAG-QS 239 Query: 735 PRKYCPEQNAQATVEALMRRVPPAVPGITFLSGGMSELEATLNLNAINRYPGKKPWALTF 914 +K PE+ A ATV+AL R VP AV G+TFLSGG SE EAT+NL+AIN P +PWALTF Sbjct: 240 AKKNTPEEIALATVQALRRTVPAAVTGVTFLSGGQSEEEATVNLSAINNVPLIRPWALTF 299 Query: 915 SFGRALQASVIQRWKGKAENIIPAQMELLKLAQANGAASLGRFEGQLETAAGGQSLFVAN 1094 S+GRALQASV++ W GK ENI Q ELLK A+ANG A+ G++ AG SLFVAN Sbjct: 300 SYGRALQASVLRAWAGKKENIAAGQNELLKRAKANGDAAQGKYVAG-SAGAGSGSLFVAN 358 Query: 1095 HAY 1103 HAY Sbjct: 359 HAY 361
>sp|P05063|ALDOC_MOUSE Fructose-bisphosphate aldolase C (Brain-type aldolase) (Aldolase 3) (Zebrin II) (Scrapie-responsive protein 2) Length = 363 Score = 462 bits (1189), Expect = e-130 Identities = 235/363 (64%), Positives = 274/363 (75%) Frame = +3 Query: 15 MPEFARYLSEHQLKELKDIANAIVSPGKGILAADESTGTLGKRFKGIHLDNTETNRRQYR 194 MP LS Q KEL DIA IV+PGKGILAADES G++ KR I ++NTE NRR YR Sbjct: 1 MPHSYPALSAEQKKELSDIALRIVTPGKGILAADESVGSMAKRLSQIGVENTEENRRLYR 60 Query: 195 QLLFATDPILARNISGVILYHETLHQKGDDGQPLVKYLQDRGIIPGIKVDTGCVPLLGTF 374 Q+LF+ D + + I GVI +HETL+QK D+G P V+ +QD+GI+ GIKVD G VPL GT Sbjct: 61 QVLFSADDRVKKCIGGVIFFHETLYQKDDNGVPFVRTIQDKGILVGIKVDKGVVPLAGTD 120 Query: 375 DECTTQGLDGLDERCAIYKKDGCHFAKWRCVLKIGQHTPSYLAMIENANVLARYAMICQK 554 E TTQGLDGL ERCA YKKDG FAKWRCVLKI TPS LA++ENANVLARYA ICQ+ Sbjct: 121 GETTTQGLDGLLERCAQYKKDGADFAKWRCVLKISDRTPSALAILENANVLARYASICQQ 180 Query: 555 NGLVPIVEPEVLPDGTHDLFTAQRVTEDVLAFVYKALADHHIYLEGTLLKPNMVTAGMQS 734 NG+VPIVEPE+LPDG HDL Q VTE VLA VYKAL+DHH+YLEGTLLKPNMVT G Sbjct: 181 NGIVPIVEPEILPDGDHDLKRCQYVTEKVLAAVYKALSDHHVYLEGTLLKPNMVTPGHAC 240 Query: 735 PRKYCPEQNAQATVEALMRRVPPAVPGITFLSGGMSELEATLNLNAINRYPGKKPWALTF 914 P KY PE+ A ATV AL R VPPAVPG+TFLSGG SE EA+LNLNAINR P +PWALTF Sbjct: 241 PIKYSPEEIAMATVTALRRTVPPAVPGVTFLSGGQSEEEASLNLNAINRCPLPRPWALTF 300 Query: 915 SFGRALQASVIQRWKGKAENIIPAQMELLKLAQANGAASLGRFEGQLETAAGGQSLFVAN 1094 S+GRALQAS + W+G+ +N A E +K A+ NG A+ GR+EG + A QSL++AN Sbjct: 301 SYGRALQASALNAWRGQRDNAGAATEEFIKRAEMNGLAAQGRYEGSGDGGAAAQSLYIAN 360 Query: 1095 HAY 1103 HAY Sbjct: 361 HAY 363
>sp|Q9GKW3|ALDOC_MACFA Fructose-bisphosphate aldolase C (Brain-type aldolase) Length = 364 Score = 461 bits (1186), Expect = e-129 Identities = 235/364 (64%), Positives = 275/364 (75%), Gaps = 1/364 (0%) Frame = +3 Query: 15 MPEFARYLSEHQLKELKDIANAIVSPGKGILAADESTGTLGKRFKGIHLDNTETNRRQYR 194 MP LS Q KEL DIA IV+PGKGILAADES G++ KR I ++NTE NRR YR Sbjct: 1 MPHSYPALSAEQKKELSDIALRIVAPGKGILAADESVGSMAKRLSQIGVENTEENRRLYR 60 Query: 195 QLLFATDPILARNISGVILYHETLHQKGDDGQPLVKYLQDRGIIPGIKVDTGCVPLLGTF 374 Q+LF+ D + + I GVI +HETL+QK D+G P V+ +QD+GI+ GIKVD G VPL GT Sbjct: 61 QVLFSADDRVKKCIGGVIFFHETLYQKDDNGVPFVRTIQDKGIVVGIKVDKGVVPLAGTD 120 Query: 375 DECTTQGLDGLDERCAIYKKDGCHFAKWRCVLKIGQHTPSYLAMIENANVLARYAMICQK 554 E TTQGLDGL ERCA YKKDG FAKWRCVLKI + TPS LA++ENANVLARYA ICQ+ Sbjct: 121 GETTTQGLDGLSERCAQYKKDGADFAKWRCVLKISERTPSALAILENANVLARYASICQQ 180 Query: 555 NGLVPIVEPEVLPDGTHDLFTAQRVTEDVLAFVYKALADHHIYLEGTLLKPNMVTAGMQS 734 NG+VPIVEPE+LPDG HDL Q VTE VLA VYKAL+DHH+YLEGTLLKPNMVT G Sbjct: 181 NGIVPIVEPEILPDGDHDLKRCQYVTEKVLAAVYKALSDHHVYLEGTLLKPNMVTPGHAC 240 Query: 735 PRKYCPEQNAQATVEALMRRVPPAVPGITFLSGGMSELEATLNLNAINRYPGKKPWALTF 914 P KY PE+ A ATV AL R VPPAVPG+TFLSGG SE EA+LNLNAINR P +PWALTF Sbjct: 241 PIKYTPEEIAMATVTALRRTVPPAVPGVTFLSGGQSEEEASLNLNAINRCPLPRPWALTF 300 Query: 915 SFGRALQASVIQRWKGKAENIIPAQMELLKLAQANGAASLGRFEGQLET-AAGGQSLFVA 1091 S+GRALQAS + W+G+ +N A E +K A+ NG A+ G++EG E A QSL++A Sbjct: 301 SYGRALQASALNAWRGQRDNAGAATEEFIKRAEVNGLAAQGKYEGSGEDGGAAAQSLYIA 360 Query: 1092 NHAY 1103 NHAY Sbjct: 361 NHAY 364
>sp|P09972|ALDOC_HUMAN Fructose-bisphosphate aldolase C (Brain-type aldolase) Length = 364 Score = 459 bits (1182), Expect = e-129 Identities = 234/364 (64%), Positives = 274/364 (75%), Gaps = 1/364 (0%) Frame = +3 Query: 15 MPEFARYLSEHQLKELKDIANAIVSPGKGILAADESTGTLGKRFKGIHLDNTETNRRQYR 194 MP LS Q KEL DIA IV+PGKGILAADES G++ KR I ++NTE NRR YR Sbjct: 1 MPHSYPALSAEQKKELSDIALRIVAPGKGILAADESVGSMAKRLSQIGVENTEENRRLYR 60 Query: 195 QLLFATDPILARNISGVILYHETLHQKGDDGQPLVKYLQDRGIIPGIKVDTGCVPLLGTF 374 Q+LF+ D + + I GVI +HETL+QK D+G P V+ +QD+GI+ GIKVD G VPL GT Sbjct: 61 QVLFSADDRVKKCIGGVIFFHETLYQKDDNGVPFVRTIQDKGIVVGIKVDKGVVPLAGTD 120 Query: 375 DECTTQGLDGLDERCAIYKKDGCHFAKWRCVLKIGQHTPSYLAMIENANVLARYAMICQK 554 E TTQGLDGL ERCA YKKDG FAKWRCVLKI + TPS LA++ENANVLARYA ICQ+ Sbjct: 121 GETTTQGLDGLSERCAQYKKDGADFAKWRCVLKISERTPSALAILENANVLARYASICQQ 180 Query: 555 NGLVPIVEPEVLPDGTHDLFTAQRVTEDVLAFVYKALADHHIYLEGTLLKPNMVTAGMQS 734 NG+VPIVEPE+LPDG HDL Q VTE VLA VYKAL+DHH+YLEGTLLKPNMVT G Sbjct: 181 NGIVPIVEPEILPDGDHDLKRCQYVTEKVLAAVYKALSDHHVYLEGTLLKPNMVTPGHAC 240 Query: 735 PRKYCPEQNAQATVEALMRRVPPAVPGITFLSGGMSELEATLNLNAINRYPGKKPWALTF 914 P KY PE+ A ATV AL R VPPAVPG+TFLSGG SE EA+ NLNAINR P +PWALTF Sbjct: 241 PIKYTPEEIAMATVTALRRTVPPAVPGVTFLSGGQSEEEASFNLNAINRCPLPRPWALTF 300 Query: 915 SFGRALQASVIQRWKGKAENIIPAQMELLKLAQANGAASLGRFEGQLET-AAGGQSLFVA 1091 S+GRALQAS + W+G+ +N A E +K A+ NG A+ G++EG E A QSL++A Sbjct: 301 SYGRALQASALNAWRGQRDNAGAATEEFIKRAEVNGLAAQGKYEGSGEDGGAAAQSLYIA 360 Query: 1092 NHAY 1103 NHAY Sbjct: 361 NHAY 364
>sp|Q5R1X4|ALDOC_PANTR Fructose-bisphosphate aldolase C (Brain-type aldolase) Length = 364 Score = 457 bits (1175), Expect = e-128 Identities = 232/364 (63%), Positives = 273/364 (75%), Gaps = 1/364 (0%) Frame = +3 Query: 15 MPEFARYLSEHQLKELKDIANAIVSPGKGILAADESTGTLGKRFKGIHLDNTETNRRQYR 194 MP LS Q KEL DIA IV+PGKGILAADES G++ KR I ++NTE NRR YR Sbjct: 1 MPHSYPALSAEQKKELSDIALRIVAPGKGILAADESVGSMAKRLSQIGVENTEENRRLYR 60 Query: 195 QLLFATDPILARNISGVILYHETLHQKGDDGQPLVKYLQDRGIIPGIKVDTGCVPLLGTF 374 Q+LF+ D + + I GVI +HETL+QK D+G P ++ + D+GI+ GIKVD G VPL GT Sbjct: 61 QVLFSADDRVKKCIGGVIFFHETLYQKDDNGVPFIRTILDKGIVVGIKVDKGVVPLAGTD 120 Query: 375 DECTTQGLDGLDERCAIYKKDGCHFAKWRCVLKIGQHTPSYLAMIENANVLARYAMICQK 554 E TTQGLDGL ERCA YKKDG FAKWRCVLKI + TPS LA++ENANVLARYA ICQ+ Sbjct: 121 GETTTQGLDGLSERCAQYKKDGADFAKWRCVLKISERTPSALAILENANVLARYASICQQ 180 Query: 555 NGLVPIVEPEVLPDGTHDLFTAQRVTEDVLAFVYKALADHHIYLEGTLLKPNMVTAGMQS 734 NG+VPIVEPE+LPDG HDL Q VTE VLA VYKAL+DHH+YLEGTLLKPNMVT G Sbjct: 181 NGIVPIVEPEILPDGDHDLKRCQYVTEKVLAAVYKALSDHHVYLEGTLLKPNMVTPGHAC 240 Query: 735 PRKYCPEQNAQATVEALMRRVPPAVPGITFLSGGMSELEATLNLNAINRYPGKKPWALTF 914 P KY PE+ A ATV AL R VPPAVPG+TFLSGG SE EA+ NLNAINR P +PWALTF Sbjct: 241 PIKYTPEEIAMATVTALRRTVPPAVPGVTFLSGGQSEEEASFNLNAINRCPLPRPWALTF 300 Query: 915 SFGRALQASVIQRWKGKAENIIPAQMELLKLAQANGAASLGRFEGQLET-AAGGQSLFVA 1091 S+GRALQAS + W+G+ +N A E +K A+ NG A+ G++EG E A QSL++A Sbjct: 301 SYGRALQASALNAWRGQRDNAEAATEEFIKRAEVNGLAAQGKYEGSGEDGGAAAQSLYIA 360 Query: 1092 NHAY 1103 NHAY Sbjct: 361 NHAY 364
>sp|P09117|ALDOC_RAT Fructose-bisphosphate aldolase C (Brain-type aldolase) Length = 363 Score = 456 bits (1174), Expect = e-128 Identities = 233/363 (64%), Positives = 273/363 (75%) Frame = +3 Query: 15 MPEFARYLSEHQLKELKDIANAIVSPGKGILAADESTGTLGKRFKGIHLDNTETNRRQYR 194 MP LS Q KEL DIA IV+PGKGILAADES G++ KR I ++NTE NRR YR Sbjct: 1 MPHSYPALSAEQKKELSDIALRIVAPGKGILAADESVGSMAKRLSQIGVENTEENRRLYR 60 Query: 195 QLLFATDPILARNISGVILYHETLHQKGDDGQPLVKYLQDRGIIPGIKVDTGCVPLLGTF 374 Q+LF+ D + + I GVI +HETL+QK D+G P V+ +Q++GI+ GIKVD G VPL GT Sbjct: 61 QVLFSADDRVKKCIGGVIFFHETLYQKDDNGVPFVRTIQEKGILVGIKVDKGVVPLAGTD 120 Query: 375 DECTTQGLDGLDERCAIYKKDGCHFAKWRCVLKIGQHTPSYLAMIENANVLARYAMICQK 554 E TTQGLDGL ERCA YKKDG FAKWRCVLKI TPS LA++ENANVLARYA ICQ+ Sbjct: 121 GETTTQGLDGLLERCAQYKKDGADFAKWRCVLKISDRTPSALAILENANVLARYASICQQ 180 Query: 555 NGLVPIVEPEVLPDGTHDLFTAQRVTEDVLAFVYKALADHHIYLEGTLLKPNMVTAGMQS 734 NG+VPIVEPE+LPDG HDL Q VTE VLA VYKAL+DHH+YLEGTLLKPNMVT G Sbjct: 181 NGIVPIVEPEILPDGDHDLKRCQFVTEKVLAAVYKALSDHHVYLEGTLLKPNMVTPGHAC 240 Query: 735 PRKYCPEQNAQATVEALMRRVPPAVPGITFLSGGMSELEATLNLNAINRYPGKKPWALTF 914 P KY PE+ A ATV AL R VPPAVPG+TFLSGG SE EA+LNLNAINR +PWALTF Sbjct: 241 PIKYSPEEIAMATVTALRRTVPPAVPGVTFLSGGQSEEEASLNLNAINRCSLPRPWALTF 300 Query: 915 SFGRALQASVIQRWKGKAENIIPAQMELLKLAQANGAASLGRFEGQLETAAGGQSLFVAN 1094 S+GRALQAS + W+G+ +N A E +K A+ NG A+ G++EG + A QSL+VAN Sbjct: 301 SYGRALQASALSAWRGQRDNAGAATEEFIKRAEMNGLAAQGKYEGSGDGGAAAQSLYVAN 360 Query: 1095 HAY 1103 HAY Sbjct: 361 HAY 363
>sp|P53445|ALF1_LAMJA Fructose-bisphosphate aldolase, muscle type Length = 363 Score = 455 bits (1170), Expect = e-127 Identities = 232/356 (65%), Positives = 273/356 (76%) Frame = +3 Query: 36 LSEHQLKELKDIANAIVSPGKGILAADESTGTLGKRFKGIHLDNTETNRRQYRQLLFATD 215 L+ Q KEL DIA IV+ GKGILAADES GT+GKR I L+NT+ +RR YRQLLF TD Sbjct: 8 LTPDQKKELADIAQRIVASGKGILAADESVGTMGKRLTQIGLENTDEHRRFYRQLLFTTD 67 Query: 216 PILARNISGVILYHETLHQKGDDGQPLVKYLQDRGIIPGIKVDTGCVPLLGTFDECTTQG 395 P + +I G+I +HET++QK D G P VK ++D GI+ GIKVD G VPL GT E TTQG Sbjct: 68 PSIKEHIGGIIFFHETMYQKTDGGVPFVKLVKDNGILVGIKVDKGVVPLAGTNGEGTTQG 127 Query: 396 LDGLDERCAIYKKDGCHFAKWRCVLKIGQHTPSYLAMIENANVLARYAMICQKNGLVPIV 575 LDGL ERCA YKKDG FAKWRCVLKI +TPS L+++ENANVLARYA ICQ+NGLVPIV Sbjct: 128 LDGLAERCAQYKKDGADFAKWRCVLKISPNTPSRLSIVENANVLARYATICQQNGLVPIV 187 Query: 576 EPEVLPDGTHDLFTAQRVTEDVLAFVYKALADHHIYLEGTLLKPNMVTAGMQSPRKYCPE 755 EPE+LPDG HDL T Q +TE VLA YKAL+DHH+YLEGTLLKPNMVT G +P PE Sbjct: 188 EPEILPDGDHDLKTCQYITEKVLAATYKALSDHHVYLEGTLLKPNMVTVGTAAPASTRPE 247 Query: 756 QNAQATVEALMRRVPPAVPGITFLSGGMSELEATLNLNAINRYPGKKPWALTFSFGRALQ 935 Q A AT+ AL R VPPAVPGITFLSGG SE +A+++LNAIN+ KPWALTFS+GRALQ Sbjct: 248 QVAMATLTALRRTVPPAVPGITFLSGGQSEEDASIHLNAINKLHLIKPWALTFSYGRALQ 307 Query: 936 ASVIQRWKGKAENIIPAQMELLKLAQANGAASLGRFEGQLETAAGGQSLFVANHAY 1103 ASV++ W GK EN+ AQ EL++ A+ NG AS G ++ AA G+SLFVANHAY Sbjct: 308 ASVLKAWGGKKENLKAAQDELMRRAKINGQASKGEYKPTGTGAAAGESLFVANHAY 363
>sp|P05065|ALDOA_RAT Fructose-bisphosphate aldolase A (Muscle-type aldolase) Length = 364 Score = 448 bits (1153), Expect = e-125 Identities = 233/365 (63%), Positives = 274/365 (75%), Gaps = 2/365 (0%) Frame = +3 Query: 15 MPEFARYLSEHQLKELKDIANAIVSPGKGILAADESTGTLGKRFKGIHLDNTETNRRQYR 194 MP L+ Q KEL DIA+ IV+PGKGILAADESTG++ KR + I +NTE NRR YR Sbjct: 1 MPHPYPALTPEQKKELADIAHRIVAPGKGILAADESTGSIAKRLQSIGTENTEENRRFYR 60 Query: 195 QLLFATDPILARNISGVILYHETLHQKGDDGQPLVKYLQDRGIIPGIKVDTGCVPLLGTF 374 QLL D + I GVIL+HETL+QK DDG+P + ++ +G + GIKVD G VPL GT Sbjct: 61 QLLLTADDRVNPCIGGVILFHETLYQKADDGRPFPQVIKSKGGVVGIKVDKGVVPLAGTN 120 Query: 375 DECTTQGLDGLDERCAIYKKDGCHFAKWRCVLKIGQHTPSYLAMIENANVLARYAMICQK 554 E TTQGLDGL ERCA YKKDG FAKWRCVLKIG+HTPS LA++ENANVLARYA ICQ+ Sbjct: 121 GETTTQGLDGLSERCAQYKKDGADFAKWRCVLKIGEHTPSSLAIMENANVLARYASICQQ 180 Query: 555 NGLVPIVEPEVLPDGTHDLFTAQRVTEDVLAFVYKALADHHIYLEGTLLKPNMVTAGMQS 734 NG+VPIVEPE+LPDG HDL Q VTE VLA VYKAL+DHH+YLEGTLLKPNMVT G Sbjct: 181 NGIVPIVEPEILPDGDHDLKRCQYVTEKVLAAVYKALSDHHVYLEGTLLKPNMVTPGHAC 240 Query: 735 PRKYCPEQNAQATVEALMRRVPPAVPGITFLSGGMSELEATLNLNAINRYPGKKPWALTF 914 +K+ E+ A ATV AL R VPPAVPG+TFLSGG SE EA++NLNAIN+ P KPWALTF Sbjct: 241 TQKFSNEEIAMATVTALRRTVPPAVPGVTFLSGGQSEEEASINLNAINKCPLLKPWALTF 300 Query: 915 SFGRALQASVIQRWKGKAENIIPAQMELLKLAQANGAASLGRF--EGQLETAAGGQSLFV 1088 S+GRALQAS ++ W GK EN+ AQ E +K A AN A G++ GQ AA +SLF+ Sbjct: 301 SYGRALQASALKAWGGKKENLKAAQEEYIKRALANSLACQGKYTPSGQ-SGAAASESLFI 359 Query: 1089 ANHAY 1103 +NHAY Sbjct: 360 SNHAY 364
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 147,127,529
Number of Sequences: 369166
Number of extensions: 3223877
Number of successful extensions: 7657
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 7287
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 7591
length of database: 68,354,980
effective HSP length: 113
effective length of database: 47,479,925
effective search space used: 13959097950
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail