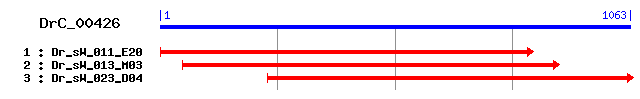
DrC_00426
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00426 (1063 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q03168|ASPP_AEDAE Lysosomal aspartic protease precursor 372 e-103 sp|Q05744|CATD_CHICK Cathepsin D precursor [Contains: Cathe... 350 4e-96 sp|P18242|CATD_MOUSE Cathepsin D precursor 334 3e-91 sp|P07339|CATD_HUMAN Cathepsin D precursor [Contains: Cathe... 334 3e-91 sp|P24268|CATD_RAT Cathepsin D precursor [Contains: Catheps... 332 8e-91 sp|P80209|CATD_BOVIN Cathepsin D precursor 331 2e-90 sp|Q9DEX3|CATD_CLUHA Cathepsin D precursor 328 2e-89 sp|Q9MZS8|CATD_SHEEP Cathepsin D precursor 323 4e-88 sp|P00795|CATD_PIG Cathepsin D [Contains: Cathepsin D light... 318 2e-86 sp|Q805F2|CATEB_XENLA Cathepsin E-B precursor 300 5e-81
>sp|Q03168|ASPP_AEDAE Lysosomal aspartic protease precursor Length = 387 Score = 372 bits (956), Expect = e-103 Identities = 188/329 (57%), Positives = 230/329 (69%), Gaps = 7/329 (2%) Frame = +2 Query: 89 RVPLKKMESARSNIKKFNMMLSHFTK--NGKVTPFPEPLNNYMDAQYFGDITIGTPPQLF 262 RV L K ESAR + + + + N P PEPL+NY+DAQY+G ITIGTPPQ F Sbjct: 22 RVQLHKTESARQHFRNVDTEIKQLRLKYNAVSGPVPEPLSNYLDAQYYGAITIGTPPQSF 81 Query: 263 TVLFDTGSSNLWVPSKKCKITNIACLTHRKYDSSKSSTYVKNGTDFSIRYGTGSLSGFLS 442 V+FDTGSSNLWVPSK+C TNIACL H KY++ KSST+ KNGT F I+YG+GSLSG+LS Sbjct: 82 KVVFDTGSSNLWVPSKECSFTNIACLMHNKYNAKKSSTFEKNGTAFHIQYGSGSLSGYLS 141 Query: 443 TDTVTIAGANVVNQTFGEAISEPGMTFVAAKFDGILGMAYPSISVDQVTPVFQNMVTQKL 622 TDTV + G +V QTF EAI+EPG+ FVAAKFDGILG+ Y SISVD V PVF NM Q L Sbjct: 142 TDTVGLGGVSVTKQTFAEAINEPGLVFVAAKFDGILGLGYSSISVDGVVPVFYNMFNQGL 201 Query: 623 VDAPIFSFYLNRDVNGKPGGEILFGGSDTNFYNGPFTYTPVTSKTYWQFKMDGLSMLGKT 802 +DAP+FSFYLNRD + GGEI+FGGSD+N Y G FTY V K YWQFKMD + + Sbjct: 202 IDAPVFSFYLNRDPSAAEGGEIIFGGSDSNKYTGDFTYLSVDRKAYWQFKMDSVKVGDTE 261 Query: 803 YCTE-CIAIADTGTSLIVGPESVIDDINSKFNGTKI-NGMTIVLCSNVAKLPVVNIQIGG 976 +C C AIADTGTSLI GP S + IN GT I NG +V CS + KLP ++ +GG Sbjct: 262 FCNNGCEAIADTGTSLIAGPVSEVTAINKAIGGTPIMNGEYMVDCSLIPKLPKISFVLGG 321 Query: 977 RNFPLNSTDYIMKVQD---TVCISGFTSL 1054 ++F L DY+++V T+C+SGF + Sbjct: 322 KSFDLEGADYVLRVAQMGKTICLSGFMGI 350
>sp|Q05744|CATD_CHICK Cathepsin D precursor [Contains: Cathepsin D light chain; Cathepsin D heavy chain] Length = 398 Score = 350 bits (898), Expect = 4e-96 Identities = 181/344 (52%), Positives = 231/344 (67%), Gaps = 19/344 (5%) Frame = +2 Query: 80 ALFRVPLKKMESAR-------SNIKKFNMMLSHFTK-----NGKVTPFPEPLNNYMDAQY 223 AL R+PL K S R S I N ++ F K P PE L NYMDAQY Sbjct: 20 ALIRIPLTKFTSTRRMLTEVGSEIPDMNA-ITQFLKFKLGFADLAEPTPEILKNYMDAQY 78 Query: 224 FGDITIGTPPQLFTVLFDTGSSNLWVPSKKCKITNIACLTHRKYDSSKSSTYVKNGTDFS 403 +G+I IGTPPQ FTV+FDTGSSNLWVPS C + +IACL H KYD+SKSSTYV+NGT+F+ Sbjct: 79 YGEIGIGTPPQKFTVVFDTGSSNLWVPSVHCHLLDIACLLHHKYDASKSSTYVENGTEFA 138 Query: 404 IRYGTGSLSGFLSTDTVTIAGANVVNQTFGEAISEPGMTFVAAKFDGILGMAYPSISVDQ 583 I YGTGSLSGFLS DTVT+ + NQ FGEA+ +PG+TF+AAKFDGILGMA+P ISVD+ Sbjct: 139 IHYGTGSLSGFLSQDTVTLGNLKIKNQIFGEAVKQPGITFIAAKFDGILGMAFPRISVDK 198 Query: 584 VTPVFQNMVTQKLVDAPIFSFYLNRDVNGKPGGEILFGGSDTNFYNGPFTYTPVTSKTYW 763 VTP F N++ QKL++ IFSFYLNRD +PGGE+L GG+D +Y+G F++ VT K YW Sbjct: 199 VTPFFDNVMQQKLIEKNIFSFYLNRDPTAQPGGELLLGGTDPKYYSGDFSWVNVTRKAYW 258 Query: 764 QFKMDGLSML-GKTYCT-ECIAIADTGTSLIVGPESVIDDINSKFNGTK--INGMTIVLC 931 Q MD + + G T C C AI DTGTSLI GP + ++ + G K I G ++ C Sbjct: 259 QVHMDSVDVANGLTLCKGGCEAIVDTGTSLITGPTKEVKELQTAI-GAKPLIKGQYVISC 317 Query: 932 SNVAKLPVVNIQIGGRNFPLNSTDYIMKVQ---DTVCISGFTSL 1054 ++ LPVV + +GG+ + L Y+ KV +T+C+SGF+ L Sbjct: 318 DKISSLPVVTLMLGGKPYQLTGEQYVFKVSAQGETICLSGFSGL 361
>sp|P18242|CATD_MOUSE Cathepsin D precursor Length = 410 Score = 334 bits (856), Expect = 3e-91 Identities = 175/352 (49%), Positives = 223/352 (63%), Gaps = 27/352 (7%) Frame = +2 Query: 80 ALFRVPLKKMESARSNIKKFNMMLSHFTKNGKVT------------PFPEPLNNYMDAQY 223 A+ R+PL+K S R + + + G +T P E L NY+DAQY Sbjct: 20 AIIRIPLRKFTSIRRTMTEVGGSVEDLILKGPITKYSMQSSPKTTEPVSELLKNYLDAQY 79 Query: 224 FGDITIGTPPQLFTVLFDTGSSNLWVPSKKCKITNIACLTHRKYDSSKSSTYVKNGTDFS 403 +GDI IGTPPQ FTV+FDTGSSNLWVPS CKI +IAC H KY+S KSSTYVKNGT F Sbjct: 80 YGDIGIGTPPQCFTVVFDTGSSNLWVPSIHCKILDIACWVHHKYNSDKSSTYVKNGTSFD 139 Query: 404 IRYGTGSLSGFLSTDTVTI---------AGANVVNQTFGEAISEPGMTFVAAKFDGILGM 556 I YG+GSLSG+LS DTV++ G V Q FGEA +PG+ FVAAKFDGILGM Sbjct: 140 IHYGSGSLSGYLSQDTVSVPCKSDQSKARGIKVEKQIFGEATKQPGIVFVAAKFDGILGM 199 Query: 557 AYPSISVDQVTPVFQNMVTQKLVDAPIFSFYLNRDVNGKPGGEILFGGSDTNFYNGPFTY 736 YP ISV+ V PVF N++ QKLVD IFSFYLNRD G+PGGE++ GG+D+ +Y+G +Y Sbjct: 200 GYPHISVNNVLPVFDNLMQQKLVDKNIFSFYLNRDPEGQPGGELMLGGTDSKYYHGELSY 259 Query: 737 TPVTSKTYWQFKMDGLSMLGK-TYCT-ECIAIADTGTSLIVGPESVIDDINSKFNGTK-I 907 VT K YWQ MD L + + T C C AI DTGTSL+VGP + ++ I Sbjct: 260 LNVTRKAYWQVHMDQLEVGNELTLCKGGCEAIVDTGTSLLVGPVEEVKELQKAIGAVPLI 319 Query: 908 NGMTIVLCSNVAKLPVVNIQIGGRNFPLNSTDYIMKVQD---TVCISGFTSL 1054 G ++ C V+ LP V +++GG+N+ L+ YI+KV T+C+SGF + Sbjct: 320 QGEYMIPCEKVSSLPTVYLKLGGKNYELHPDKYILKVSQGGKTICLSGFMGM 371
>sp|P07339|CATD_HUMAN Cathepsin D precursor [Contains: Cathepsin D light chain; Cathepsin D heavy chain] Length = 412 Score = 334 bits (856), Expect = 3e-91 Identities = 173/354 (48%), Positives = 224/354 (63%), Gaps = 29/354 (8%) Frame = +2 Query: 80 ALFRVPLKKMESARSNIKKFNMMLSHFTKNGKVT------------PFPEPLNNYMDAQY 223 AL R+PL K S R + + + G V+ P PE L NYMDAQY Sbjct: 20 ALVRIPLHKFTSIRRTMSEVGGSVEDLIAKGPVSKYSQAVPAVTEGPIPEVLKNYMDAQY 79 Query: 224 FGDITIGTPPQLFTVLFDTGSSNLWVPSKKCKITNIACLTHRKYDSSKSSTYVKNGTDFS 403 +G+I IGTPPQ FTV+FDTGSSNLWVPS CK+ +IAC H KY+S KSSTYVKNGT F Sbjct: 80 YGEIGIGTPPQCFTVVFDTGSSNLWVPSIHCKLLDIACWIHHKYNSDKSSTYVKNGTSFD 139 Query: 404 IRYGTGSLSGFLSTDTVTI-----------AGANVVNQTFGEAISEPGMTFVAAKFDGIL 550 I YG+GSLSG+LS DTV++ G V Q FGEA +PG+TF+AAKFDGIL Sbjct: 140 IHYGSGSLSGYLSQDTVSVPCQSASSASALGGVKVERQVFGEATKQPGITFIAAKFDGIL 199 Query: 551 GMAYPSISVDQVTPVFQNMVTQKLVDAPIFSFYLNRDVNGKPGGEILFGGSDTNFYNGPF 730 GMAYP ISV+ V PVF N++ QKLVD IFSFYL+RD + +PGGE++ GG+D+ +Y G Sbjct: 200 GMAYPRISVNNVLPVFDNLMQQKLVDQNIFSFYLSRDPDAQPGGELMLGGTDSKYYKGSL 259 Query: 731 TYTPVTSKTYWQFKMDGLSML-GKTYCTE-CIAIADTGTSLIVGPESVIDDINSKFNGTK 904 +Y VT K YWQ +D + + G T C E C AI DTGTSL+VGP + ++ Sbjct: 260 SYLNVTRKAYWQVHLDQVEVASGLTLCKEGCEAIVDTGTSLMVGPVDEVRELQKAIGAVP 319 Query: 905 -INGMTIVLCSNVAKLPVVNIQIGGRNFPLNSTDYIMKVQD---TVCISGFTSL 1054 I G ++ C V+ LP + +++GG+ + L+ DY +KV T+C+SGF + Sbjct: 320 LIQGEYMIPCEKVSTLPAITLKLGGKGYKLSPEDYTLKVSQAGKTLCLSGFMGM 373
>sp|P24268|CATD_RAT Cathepsin D precursor [Contains: Cathepsin D 12 kDa light chain; Cathepsin D 9 kDa light chain; Cathepsin D 34 kDa heavy chain; Cathepsin D 30 kDa heavy chain] Length = 407 Score = 332 bits (852), Expect = 8e-91 Identities = 171/349 (48%), Positives = 224/349 (64%), Gaps = 24/349 (6%) Frame = +2 Query: 80 ALFRVPLKKMESARSNIKKFNMMLSHFTKNGKVT------------PFPEPLNNYMDAQY 223 AL R+PL+K S R + + + G +T P E L NY+DAQY Sbjct: 20 ALIRIPLRKFTSIRRTMTEVGGSVEDLILKGPITKYSMQSSPRTKEPVSELLKNYLDAQY 79 Query: 224 FGDITIGTPPQLFTVLFDTGSSNLWVPSKKCKITNIACLTHRKYDSSKSSTYVKNGTDFS 403 +G+I IGTPPQ FTV+FDTGSSNLWVPS CK+ +IAC H KY+S KSSTYVKNGT F Sbjct: 80 YGEIGIGTPPQCFTVVFDTGSSNLWVPSIHCKLLDIACWVHHKYNSDKSSTYVKNGTSFD 139 Query: 404 IRYGTGSLSGFLSTDTVTI------AGANVVNQTFGEAISEPGMTFVAAKFDGILGMAYP 565 I YG+GSLSG+LS DTV++ G V Q FGEA +PG+ F+AAKFDGILGM YP Sbjct: 140 IHYGSGSLSGYLSQDTVSVPCKSDLGGIKVEKQIFGEATKQPGVVFIAAKFDGILGMGYP 199 Query: 566 SISVDQVTPVFQNMVTQKLVDAPIFSFYLNRDVNGKPGGEILFGGSDTNFYNGPFTYTPV 745 ISV++V PVF N++ QKLV+ IFSFYLNRD G+PGGE++ GG+D+ +Y+G +Y V Sbjct: 200 FISVNKVLPVFDNLMKQKLVEKNIFSFYLNRDPTGQPGGELMLGGTDSRYYHGELSYLNV 259 Query: 746 TSKTYWQFKMDGLSMLGK-TYCT-ECIAIADTGTSLIVGPESVIDDINSKFNGTK-INGM 916 T K YWQ MD L + + T C C AI DTGTSL+VGP + ++ I G Sbjct: 260 TRKAYWQVHMDQLEVGSELTLCKGGCEAIVDTGTSLLVGPVDEVKELQKAIGAVPLIQGE 319 Query: 917 TIVLCSNVAKLPVVNIQIGGRNFPLNSTDYIMKVQD---TVCISGFTSL 1054 ++ C V+ LP++ ++GG+N+ L+ YI+KV T+C+SGF + Sbjct: 320 YMIPCEKVSSLPIITFKLGGQNYELHPEKYILKVSQAGKTICLSGFMGM 368
>sp|P80209|CATD_BOVIN Cathepsin D precursor Length = 390 Score = 331 bits (848), Expect = 2e-90 Identities = 172/351 (49%), Positives = 224/351 (63%), Gaps = 27/351 (7%) Frame = +2 Query: 83 LFRVPLKKMESARSNIKKFNMMLSHFTKNGKVT------------PFPEPLNNYMDAQYF 226 + R+PL K S R + + + G ++ P PE L NYMDAQY+ Sbjct: 1 VIRIPLHKFTSIRRTMSEAAGXVXXLIAKGPISKYATGEPAVRQGPIPELLKNYMDAQYY 60 Query: 227 GDITIGTPPQLFTVLFDTGSSNLWVPSKKCKITNIACLTHRKYDSSKSSTYVKNGTDFSI 406 G+I IGTPPQ FTV+FDTGS+NLWVPS CK+ +IAC THRKY+S KSSTYVKNGT F I Sbjct: 61 GEIGIGTPPQCFTVVFDTGSANLWVPSIHCKLLDIACWTHRKYNSDKSSTYVKNGTTFDI 120 Query: 407 RYGTGSLSGFLSTDTVTI---------AGANVVNQTFGEAISEPGMTFVAAKFDGILGMA 559 YG+GSLSG+LS DTV++ G V QTFGEAI +PG+ F+AAKFDGILGMA Sbjct: 121 HYGSGSLSGYLSQDTVSVPCNPSSSSPGGVTVQRQTFGEAIKQPGVVFIAAKFDGILGMA 180 Query: 560 YPSISVDQVTPVFQNMVTQKLVDAPIFSFYLNRDVNGKPGGEILFGGSDTNFYNGPFTYT 739 YP ISV+ V PVF N++ QKLVD +FSF+LNRD +PGGE++ GG+D+ +Y G + Sbjct: 181 YPRISVNNVLPVFDNLMQQKLVDKNVFSFFLNRDPKAQPGGELMLGGTDSKYYRGSLMFH 240 Query: 740 PVTSKTYWQFKMDGLSM-LGKTYCT-ECIAIADTGTSLIVGPESVIDDINSKFNGTK-IN 910 VT + YWQ MD L + T C C AI DTGTSLIVGP + ++ I Sbjct: 241 NVTRQAYWQIHMDQLDVGSSLTVCKGGCEAIVDTGTSLIVGPVEEVRELQKAIGAVPLIQ 300 Query: 911 GMTIVLCSNVAKLPVVNIQIGGRNFPLNSTDYIMKV---QDTVCISGFTSL 1054 G ++ C V+ LP V +++GG+++ L+ DY +KV + TVC+SGF + Sbjct: 301 GEYMIPCEKVSSLPEVTVKLGGKDYALSPEDYALKVSQAETTVCLSGFMGM 351
>sp|Q9DEX3|CATD_CLUHA Cathepsin D precursor Length = 396 Score = 328 bits (840), Expect = 2e-89 Identities = 166/342 (48%), Positives = 218/342 (63%), Gaps = 17/342 (4%) Frame = +2 Query: 80 ALFRVPLKKMESARSNIKKFNMMLSHFTKN-----------GKVTPFPEPLNNYMDAQYF 226 A+ R+PLKK S R + + + P PE L NYMDAQY+ Sbjct: 18 AIVRIPLKKFRSIRRTLSDSGLNVEQLLAGTNSLQHNQGFPSSNAPTPETLKNYMDAQYY 77 Query: 227 GDITIGTPPQLFTVLFDTGSSNLWVPSKKCKITNIACLTHRKYDSSKSSTYVKNGTDFSI 406 G+I +GTP Q+FTV+FDTGSSNLW+PS C T+IACL H KY+ +KSSTYVKNGT+F+I Sbjct: 78 GEIGLGTPVQMFTVVFDTGSSNLWLPSIHCSFTDIACLLHHKYNGAKSSTYVKNGTEFAI 137 Query: 407 RYGTGSLSGFLSTDTVTIAGANVVNQTFGEAISEPGMTFVAAKFDGILGMAYPSISVDQV 586 +YG+GSLSG+LS D+ TI V Q FGEAI +PG+ F+AAKFDGILGMAYP ISVD V Sbjct: 138 QYGSGSLSGYLSQDSCTIGDIVVEKQLFGEAIKQPGVAFIAAKFDGILGMAYPRISVDGV 197 Query: 587 TPVFQNMVTQKLVDAPIFSFYLNRDVNGKPGGEILFGGSDTNFYNGPFTYTPVTSKTYWQ 766 PVF M++QK V+ +FSFYLNR+ + +PGGE+L GG+D +Y G F Y PVT + YWQ Sbjct: 198 PPVFDMMMSQKKVEQNVFSFYLNRNPDTEPGGELLLGGTDPKYYTGDFNYVPVTRQAYWQ 257 Query: 767 FKMDGLSMLGK-TYCTE-CIAIADTGTSLIVGPESVIDDINSKFNGTK-INGMTIVLCSN 937 MDG+S+ + T C + C AI DTGTSLI GP + + + I G ++ C Sbjct: 258 IHMDGMSIGSQLTLCKDGCEAIVDTGTSLITGPPAEVRALQKAIGAIPLIQGEYMIDCKK 317 Query: 938 VAKLPVVNIQIGGRNFPLNSTDYIMKVQD---TVCISGFTSL 1054 V LP ++ +GG+ + L Y++K T+C+SG L Sbjct: 318 VPTLPTISFNVGGKTYSLTGEQYVLKESQGGKTICLSGLMGL 359
>sp|Q9MZS8|CATD_SHEEP Cathepsin D precursor Length = 365 Score = 323 bits (829), Expect = 4e-88 Identities = 163/306 (53%), Positives = 208/306 (67%), Gaps = 15/306 (4%) Frame = +2 Query: 182 PFPEPLNNYMDAQYFGDITIGTPPQLFTVLFDTGSSNLWVPSKKCKITNIACLTHRKYDS 361 P PE L NYMDAQY+G+I IGTPPQ FTV+FDTGS+NLWVPS CK+ +IAC H KY+S Sbjct: 41 PIPELLTNYMDAQYYGEIGIGTPPQCFTVVFDTGSANLWVPSIHCKLLDIACWVHHKYNS 100 Query: 362 SKSSTYVKNGTDFSIRYGTGSLSGFLSTDTVTI---------AGANVVNQTFGEAISEPG 514 KSSTYVKNGT F I YG+GSLSG+LS DTV++ G V QTFGEAI +PG Sbjct: 101 DKSSTYVKNGTTFDIHYGSGSLSGYLSQDTVSVPCNPSSSSPGGVTVQRQTFGEAIKQPG 160 Query: 515 MTFVAAKFDGILGMAYPSISVDQVTPVFQNMVTQKLVDAPIFSFYLNRDVNGKPGGEILF 694 + F+AAKFDGILGMAYP ISV+ V PVF N++ QKLVD +FSF+LNRD +PG E++ Sbjct: 161 VVFIAAKFDGILGMAYPRISVNNVLPVFDNLMRQKLVDKNVFSFFLNRDPKAQPGEELML 220 Query: 695 GGSDTNFYNGPFTYTPVTSKTYWQFKMDGLSM-LGKTYCT-ECIAIADTGTSLIVGPESV 868 GG+D+ +Y G TY VT + YWQ MD L + T C C AI DTGTSL+VGP Sbjct: 221 GGTDSKYYRGSLTYHNVTRQAYWQIHMDQLDVGSSLTVCKGGCEAIVDTGTSLMVGPVDE 280 Query: 869 IDDINSKFNGTK-INGMTIVLCSNVAKLPVVNIQIGGRNFPLNSTDYIMKVQD---TVCI 1036 + +++ I G ++ C V+ LP V +++GG+++ L+ DY +KV TVC+ Sbjct: 281 VRELHKAIGAVPLIQGEYMIPCEKVSSLPQVTLKLGGKDYTLSPEDYTLKVSQAGTTVCL 340 Query: 1037 SGFTSL 1054 SGF + Sbjct: 341 SGFMGM 346
>sp|P00795|CATD_PIG Cathepsin D [Contains: Cathepsin D light chain; Cathepsin D heavy chain] Length = 345 Score = 318 bits (815), Expect = 2e-86 Identities = 164/307 (53%), Positives = 206/307 (67%), Gaps = 16/307 (5%) Frame = +2 Query: 182 PFPEPLNNYMDAQYFGDITIGTPPQLFTVLFDTGSSNLWVPSKKCKITNIACLTHRKYDS 361 P PE L NYMDAQY+G+I IGTPPQ FTV+FDTGSSNLWVPS CK+ +IAC H KY+S Sbjct: 2 PIPEVLKNYMDAQYYGEIGIGTPPQCFTVVFDTGSSNLWVPSIHCKLLDIACWIHHKYNS 61 Query: 362 SKSSTYVKNGTDFSIRYGTGSLSGFLST-DTVT---------IAGANVVNQTFGEAISEP 511 KSSTYVKNGT F+I YG+GSLSG+LS+ DTV+ + G V QTFGEA +P Sbjct: 62 GKSSTYVKNGTTFAIHYGSGSLSGYLSSQDTVSVPCNSALSGVGGIKVERQTFGEATKQP 121 Query: 512 GMTFVAAKFDGILGMAYPSISVDQVTPVFQNMVTQKLVDAPIFSFYLNRDVNGKPGGEIL 691 G+TF+AAKFDGILGMAYP ISV+ V PVF N++ QKLVD IFSFYLNRD +PGGE++ Sbjct: 122 GLTFIAAKFDGILGMAYPRISVNNVVPVFDNLMQQKLVDKDIFSFYLNRDPGAQPGGELM 181 Query: 692 FGGSDTNFYNGPFTYTPVTSKTYWQFKMDGLSM-LGKTYCT-ECIAIADTGTSLIVGPES 865 GG D+ +Y G Y VT K YWQ M+ +++ T C C AI DTGTSLIVG Sbjct: 182 LGGIDSKYYKGSLDYHNVTRKAYWQIHMNQVAVGSSLTLCKGGCEAIVDTGTSLIVGQPE 241 Query: 866 VIDDINSKFNGTK-INGMTIVLCSNVAKLPVVNIQIGGRNFPLNSTDYIMKVQ---DTVC 1033 + ++ I G ++ C V LP V + +GG+ + L+S +Y +KV T+C Sbjct: 242 EVRELGKAIGAVPLIQGEYMIPCEKVPSLPDVTVTLGGKKYKLSSENYTLKVSQAGQTIC 301 Query: 1034 ISGFTSL 1054 +SGF + Sbjct: 302 LSGFMGM 308
>sp|Q805F2|CATEB_XENLA Cathepsin E-B precursor Length = 397 Score = 300 bits (768), Expect = 5e-81 Identities = 159/342 (46%), Positives = 212/342 (61%), Gaps = 14/342 (4%) Frame = +2 Query: 71 LVVALFRVPLKKMESARSNIKKFNMMLSHFTKNG------------KVTPFPEPLNNYMD 214 LV L RVPLK+ +S R K+ + +T+ G P EPL NYMD Sbjct: 13 LVYGLIRVPLKRQKSIRKTPKEKGKLSHVWTQQGIDMVQYTDSCNNDQAP-SEPLINYMD 71 Query: 215 AQYFGDITIGTPPQLFTVLFDTGSSNLWVPSKKCKITNIACLTHRKYDSSKSSTYVKNGT 394 QYFG+I+IGTPPQ FTV+FDTGSSNLWVPS C + AC H ++ SSTY NG Sbjct: 72 VQYFGEISIGTPPQNFTVIFDTGSSNLWVPSVYC--ISPACAQHNRFQPQLSSTYESNGN 129 Query: 395 DFSIRYGTGSLSGFLSTDTVTIAGANVVNQTFGEAISEPGMTFVAAKFDGILGMAYPSIS 574 +FS++YGTGSLSG + D+VT+ G V NQ FGE++SEPG TFV A FDGILG+ YPSI+ Sbjct: 130 NFSLQYGTGSLSGVIGIDSVTVEGILVQNQQFGESVSEPGSTFVDASFDGILGLGYPSIA 189 Query: 575 VDQVTPVFQNMVTQKLVDAPIFSFYLNRDVNGKPGGEILFGGSDTNFYNGPFTYTPVTSK 754 V TPVF NM+ Q LV+ P+FS Y++RD N GGE++FGG D + ++G + PVT++ Sbjct: 190 VGGCTPVFDNMIAQNLVELPMFSVYMSRDPNSPVGGELVFGGFDASRFSGQLNWVPVTNQ 249 Query: 755 TYWQFKMDGLSMLGK-TYCT-ECIAIADTGTSLIVGPESVIDDINSKFNGTKINGMTIVL 928 YWQ ++D + + G+ +C+ C AI DTGTS+I GP S I + S + NG V Sbjct: 250 GYWQIQLDNIQINGEVVFCSGGCQAIVDTGTSMITGPSSDIVQLQSIIGASAANGDYEVD 309 Query: 929 CSNVAKLPVVNIQIGGRNFPLNSTDYIMKVQDTVCISGFTSL 1054 C+ + K+P + I G + + Y ++ D VC SGF L Sbjct: 310 CTVLNKMPTMTFTINGIGYQMTPQQYTLQDDDGVCSSGFQGL 351
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 123,932,146
Number of Sequences: 369166
Number of extensions: 2655700
Number of successful extensions: 7046
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 6451
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 6680
length of database: 68,354,980
effective HSP length: 112
effective length of database: 47,664,660
effective search space used: 11487183060
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail