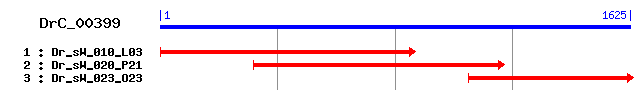
DrC_00399
- Cluster detail
- GO Category :
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00399 (1625 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q13823|NOG2_HUMAN Nucleolar GTP-binding protein 2 (Autoa... 335 2e-91 sp|Q6CSP9|NOG2_KLULA Nucleolar GTP-binding protein 2 332 1e-90 sp|Q99LH1|NOG2_MOUSE Nucleolar GTP-binding protein 2 332 2e-90 sp|Q75DA4|NOG2_ASHGO Nucleolar GTP-binding protein 2 327 6e-89 sp|O14236|NOG2_SCHPO Nucleolar GTP-binding protein 2 321 3e-87 sp|Q6C036|NOG2_YARLI Nucleolar GTP-binding protein 2 321 4e-87 sp|Q9C3Z4|NOG2_PNECA Nucleolar GTP-binding protein 2 (Bindi... 318 4e-86 sp|Q6FWS1|NOG2_CANGA Nucleolar GTP-binding protein 2 315 2e-85 sp|P53742|NOG2_YEAST Nucleolar GTP-binding protein 2 314 5e-85 sp|Q7SHR8|NOG2_NEUCR Nucleolar GTP-binding protein 2 303 7e-82
>sp|Q13823|NOG2_HUMAN Nucleolar GTP-binding protein 2 (Autoantigen NGP-1) Length = 731 Score = 335 bits (859), Expect = 2e-91 Identities = 178/390 (45%), Positives = 255/390 (65%), Gaps = 12/390 (3%) Frame = +1 Query: 1 KWFGNTRVISEKDLMKFQEYQNTY--DPFQVTLRKTVMPLSL-----NLHKTKKNKPNFK 159 KWFGNTRVI + L KFQE +T DP++V ++++ +P+SL H K + + + Sbjct: 79 KWFGNTRVIKQSSLQKFQEEMDTVMKDPYKVVMKQSKLPMSLLHDRIRPHNLKVHILDTE 138 Query: 160 TLQPYFKRKITVKESNIKARTFEELSAQTKTALEEFNAKQSMLSIDDD---KDEATHGVF 330 + + F K K N+ A + L + + E ++ + + +D ++EA ++ Sbjct: 139 SFETTFGPKSQRKRPNLFASDMQSLIENAEMSTESYDQGKDRDLVTEDTGVRNEAQEEIY 198 Query: 331 RAGTSNRIWQEVYKVIDSSDVLLYVLDARNPMGMRSPKIELDIRNKQ--KKIIFVLNKVD 504 + G S RIW E+YKVIDSSDV++ VLDAR+PMG RSP IE ++ ++ K +IFVLNK D Sbjct: 199 KKGQSKRIWGELYKVIDSSDVVVQVLDARDPMGTRSPHIETYLKKEKPWKHLIFVLNKCD 258 Query: 505 LVPSGVTRHWEKLLNKEYPTIMFYADVKHPLGKDRLICLLRQFRDKMKDRRQISVGIIGF 684 LVP+ T+ W +L+++YPT+ F+A + +P GK I LLRQF D++QISVG IG+ Sbjct: 259 LVPTWATKRWVAVLSQDYPTLAFHASLTNPFGKGAFIQLLRQFGKLHTDKKQISVGFIGY 318 Query: 685 PNTGKSSLINTLKGEKVCKVAPIAGETKVWQYISLMKSIDLIDCPGTVTSEKDEKLSETD 864 PN GKSS+INTL+ +KVC VAPIAGETKVWQYI+LM+ I LIDCPG V +D SETD Sbjct: 319 PNVGKSSVINTLRSKKVCNVAPIAGETKVWQYITLMRRIFLIDCPGVVYPSED---SETD 375 Query: 865 LILRNSVRVEKVEDADRHIPAVLNIVNRDVINKLYKINQPWNECEQFLEMVAKSRGRLLK 1044 ++L+ V+VEK++ + HI AVL + I+K YKI+ W E FLE +A G+LLK Sbjct: 376 IVLKGVVQVEKIKSPEDHIGAVLERAKPEYISKTYKIDS-WENAEDFLEKLAFRTGKLLK 434 Query: 1045 GGQPDIMTVAKEILKDFNTGKLVWYVKPEN 1134 GG+PD+ TV K +L D+ G++ ++VKP N Sbjct: 435 GGEPDLQTVGKMVLNDWQRGRIPFFVKPPN 464
>sp|Q6CSP9|NOG2_KLULA Nucleolar GTP-binding protein 2 Length = 513 Score = 332 bits (852), Expect = 1e-90 Identities = 191/404 (47%), Positives = 253/404 (62%), Gaps = 20/404 (4%) Frame = +1 Query: 1 KWFGNTRVISEKDLMKFQEY--QNTYDPFQVTLRKTVMPLSLNLHKTKKNKPNFKTLQ-- 168 +WFGNTRVIS+ L F+E +N D +QV LR+ +P+SL K P K L+ Sbjct: 76 RWFGNTRVISQDSLSHFREALGENKRDSYQVLLRRNKLPMSLLNEKDSAESPTAKILETE 135 Query: 169 PY---FKRKITVKESNIKARTFEELSAQTKTALEEFNAKQ---SMLSIDDDKDE------ 312 P+ F K K+ I A + EEL + T T + F KQ S L + ++E Sbjct: 136 PFEQTFGPKAQRKKPRIAASSLEELISSTSTDNKTFEEKQELDSTLGLMGKQEEEDGWTQ 195 Query: 313 -ATHGVFRAGTSNRIWQEVYKVIDSSDVLLYVLDARNPMGMRSPKIELDIRNK--QKKII 483 A +F G S RIW E+YKVIDSSDV+++VLDAR+P+G R + + N+ K +I Sbjct: 196 AAKEAIFHKGQSKRIWNELYKVIDSSDVVIHVLDARDPLGTRCKSVTDYMTNETPHKHLI 255 Query: 484 FVLNKVDLVPSGVTRHWEKLLNKEYPTIMFYADVKHPLGKDRLICLLRQFRDKMKDRRQI 663 +VLNK DLVP+ V W K L+KE PT+ F+A + + GK LI LLRQF KDR QI Sbjct: 256 YVLNKCDLVPTWVAAAWVKHLSKERPTLAFHASITNSFGKGSLIQLLRQFSQLHKDRHQI 315 Query: 664 SVGIIGFPNTGKSSLINTLKGEKVCKVAPIAGETKVWQYISLMKSIDLIDCPGTV-TSEK 840 SVG IG+PNTGKSS+INTL+ +KVC+VAPI GETKVWQYI+LMK I LIDCPG V S K Sbjct: 316 SVGFIGYPNTGKSSIINTLRKKKVCQVAPIPGETKVWQYITLMKRIFLIDCPGIVPPSSK 375 Query: 841 DEKLSETDLILRNSVRVEKVEDADRHIPAVLNIVNRDVINKLYKINQPWNECEQFLEMVA 1020 D SE D++ R VRVE V +++IP +L R + + Y+I+ W + F+EM+A Sbjct: 376 D---SEEDILFRGVVRVEHVSHPEQYIPGILKRCKRQHLERTYEIS-GWKDSVDFIEMIA 431 Query: 1021 KSRGRLLKGGQPDIMTVAKEILKDFNTGKLVWYVKPENYDNSKE 1152 + +GRLLKGG+PD V+K+IL DFN GK+ W+V P D +E Sbjct: 432 RKQGRLLKGGEPDESGVSKQILNDFNRGKIPWFVPPPEKDKIEE 475
>sp|Q99LH1|NOG2_MOUSE Nucleolar GTP-binding protein 2 Length = 728 Score = 332 bits (851), Expect = 2e-90 Identities = 179/390 (45%), Positives = 256/390 (65%), Gaps = 12/390 (3%) Frame = +1 Query: 1 KWFGNTRVISEKDLMKFQEYQNTY--DPFQVTLRKTVMPLSL-----NLHKTKKNKPNFK 159 KWFGNTRVI + L KFQE + DP++V ++++ +P+SL H K + + + Sbjct: 79 KWFGNTRVIKQASLQKFQEEMDKVMKDPYKVVMKQSKLPMSLLHDRIQPHNAKVHILDTE 138 Query: 160 TLQPYFKRKITVKESNIKARTFEELSAQTKTALEEFN-AKQSMLSIDDD--KDEATHGVF 330 + + F K K N+ A + L + + E ++ K L ++D ++EA ++ Sbjct: 139 SFESTFGPKSQRKRPNLFASDMQSLLENAEMSTESYDQGKDRDLVMEDTGVRNEAQEEIY 198 Query: 331 RAGTSNRIWQEVYKVIDSSDVLLYVLDARNPMGMRSPKIELDIRNKQ--KKIIFVLNKVD 504 + G S RIW E+YKVIDSSDV++ VLDAR+PMG RSP IE ++ ++ K +IFVLNK D Sbjct: 199 KKGQSKRIWGELYKVIDSSDVVVQVLDARDPMGTRSPHIEAYLKKEKPWKHLIFVLNKCD 258 Query: 505 LVPSGVTRHWEKLLNKEYPTIMFYADVKHPLGKDRLICLLRQFRDKMKDRRQISVGIIGF 684 LVP+ T+ W +L+++YPT+ F+A + +P GK I LLRQF D++QISVG IG+ Sbjct: 259 LVPTWATKRWVAVLSQDYPTLAFHASLTNPFGKGAFIQLLRQFGKLHTDKKQISVGFIGY 318 Query: 685 PNTGKSSLINTLKGEKVCKVAPIAGETKVWQYISLMKSIDLIDCPGTVTSEKDEKLSETD 864 PN GKSS+INTL+ +KVC VAPIAGETKVWQYI+LM+ I LIDCPG V +D SETD Sbjct: 319 PNVGKSSVINTLRSKKVCNVAPIAGETKVWQYITLMRRIFLIDCPGVVYPSED---SETD 375 Query: 865 LILRNSVRVEKVEDADRHIPAVLNIVNRDVINKLYKINQPWNECEQFLEMVAKSRGRLLK 1044 ++L+ V+VEK++ HI AVL + I+K YKI + W E FLE +A G+LLK Sbjct: 376 IVLKGVVQVEKIKAPQDHIGAVLERAKPEYISKTYKI-ESWENAEDFLEKLALRTGKLLK 434 Query: 1045 GGQPDIMTVAKEILKDFNTGKLVWYVKPEN 1134 GG+PD++TV+K +L D+ G++ ++VKP N Sbjct: 435 GGEPDMLTVSKMVLNDWQRGRIPFFVKPPN 464
>sp|Q75DA4|NOG2_ASHGO Nucleolar GTP-binding protein 2 Length = 502 Score = 327 bits (838), Expect = 6e-89 Identities = 184/407 (45%), Positives = 255/407 (62%), Gaps = 20/407 (4%) Frame = +1 Query: 1 KWFGNTRVISEKDLMKFQEY--QNTYDPFQVTLRKTVMPLSLNLHKTKKNKPNFKTL--Q 168 +WFGNTRVIS+ L F++ D +QV LR+ +P+SL K P K + + Sbjct: 76 RWFGNTRVISQDALQHFRDALGDKKNDSYQVLLRRNKLPMSLLEEKDTSESPTAKIIDTE 135 Query: 169 PY---FKRKITVKESNIKARTFEELSAQTKTALEEFNAKQSMLSI----------DDDKD 309 PY F K K+ + A + E+L+ T + +++ K+ + S D Sbjct: 136 PYGATFGPKAQRKKPRVAAASLEDLAKATDSDSQKYEEKKELDSTLGLMAATEQEDGWSQ 195 Query: 310 EATHGVFRAGTSNRIWQEVYKVIDSSDVLLYVLDARNPMGMRSPKIELDIRNK--QKKII 483 A +F G S RIW E+YKVIDSSDV+++VLDAR+P+G R +E ++ + K +I Sbjct: 196 VAKEAIFHKGQSKRIWNELYKVIDSSDVVIHVLDARDPLGTRCKSVEEYMKKETPHKHLI 255 Query: 484 FVLNKVDLVPSGVTRHWEKLLNKEYPTIMFYADVKHPLGKDRLICLLRQFRDKMKDRRQI 663 +VLNK DLVP+ + W K L+K+ PT+ F+A + + GK LI LLRQF KDR+QI Sbjct: 256 YVLNKCDLVPTWLAAAWVKHLSKDRPTLAFHASITNSFGKGSLIQLLRQFSQLHKDRQQI 315 Query: 664 SVGIIGFPNTGKSSLINTLKGEKVCKVAPIAGETKVWQYISLMKSIDLIDCPGTV-TSEK 840 SVG IG+PNTGKSS+INTL+ +KVC+VAPI GETKVWQYI+LMK I LIDCPG V S K Sbjct: 316 SVGFIGYPNTGKSSIINTLRKKKVCQVAPIPGETKVWQYITLMKRIFLIDCPGIVPPSAK 375 Query: 841 DEKLSETDLILRNSVRVEKVEDADRHIPAVLNIVNRDVINKLYKINQPWNECEQFLEMVA 1020 D +E D++ R VRVE V +++IPAVL R + + Y+I+ W + +F+EM+A Sbjct: 376 D---TEEDILFRGVVRVEHVSHPEQYIPAVLRRCKRHHLERTYEIS-GWADATEFIEMLA 431 Query: 1021 KSRGRLLKGGQPDIMTVAKEILKDFNTGKLVWYVKPENYDNSKETLK 1161 + +GRLLKGG+PD VAK++L DFN GK+ W+V P + D ET K Sbjct: 432 RKQGRLLKGGEPDETGVAKQVLNDFNRGKIPWFVSPPDRDPQPETSK 478
>sp|O14236|NOG2_SCHPO Nucleolar GTP-binding protein 2 Length = 537 Score = 321 bits (823), Expect = 3e-87 Identities = 178/399 (44%), Positives = 250/399 (62%), Gaps = 13/399 (3%) Frame = +1 Query: 1 KWFGNTRVISEKDLMKFQEY--QNTYDPFQVTLRKTVMPLSLNLHKTKKNKPNFKTLQPY 174 +WF NTRVI++ L +F+E Q DP+QV LR+ +P+SL T+ K +P+ Sbjct: 78 RWFNNTRVIAQPTLTQFREAMGQKLNDPYQVLLRRNKLPMSLLQENTEIPKVRVLESEPF 137 Query: 175 ---FKRKITVKESNIKARTFEELSAQTKTALEEFNAK--QSMLSIDDDKDE----ATHGV 327 F K K I + EL+ ++ + K + +L+ D+ D+ A + Sbjct: 138 ENTFGPKSQRKRPKISFDSVAELAKESDEKQNAYEEKIEERILANPDESDDVMLAARDAI 197 Query: 328 FRAGTSNRIWQEVYKVIDSSDVLLYVLDARNPMGMRSPKIELDIRNK--QKKIIFVLNKV 501 F G S RIW E+YKVIDSSDVL+ VLDAR+P+G R +E +RN+ K +I VLNKV Sbjct: 198 FSKGQSKRIWNELYKVIDSSDVLIQVLDARDPVGTRCGTVERYLRNEASHKHMILVLNKV 257 Query: 502 DLVPSGVTRHWEKLLNKEYPTIMFYADVKHPLGKDRLICLLRQFRDKMKDRRQISVGIIG 681 DLVP+ V W K+L KEYPTI F+A + + GK LI +LRQF D++QISVG+IG Sbjct: 258 DLVPTSVAAAWVKILAKEYPTIAFHASINNSFGKGSLIQILRQFASLHSDKKQISVGLIG 317 Query: 682 FPNTGKSSLINTLKGEKVCKVAPIAGETKVWQYISLMKSIDLIDCPGTVTSEKDEKLSET 861 FPN GKSS+INTL+ +KVC VAPI GETKVWQY++LMK I LIDCPG V ++ S+ Sbjct: 318 FPNAGKSSIINTLRKKKVCNVAPIPGETKVWQYVALMKRIFLIDCPGIVPPSSND--SDA 375 Query: 862 DLILRNSVRVEKVEDADRHIPAVLNIVNRDVINKLYKINQPWNECEQFLEMVAKSRGRLL 1041 +L+L+ VRVE V + + +IP VL+ + + Y+I+ WN+ +FL +AK GRLL Sbjct: 376 ELLLKGVVRVENVSNPEAYIPTVLSRCKVKHLERTYEIS-GWNDSTEFLAKLAKKGGRLL 434 Query: 1042 KGGQPDIMTVAKEILKDFNTGKLVWYVKPENYDNSKETL 1158 KGG+PD +VAK +L DF GK+ W++ P+ +S + + Sbjct: 435 KGGEPDEASVAKMVLNDFMRGKIPWFIGPKGLSSSNDEI 473
>sp|Q6C036|NOG2_YARLI Nucleolar GTP-binding protein 2 Length = 509 Score = 321 bits (822), Expect = 4e-87 Identities = 181/397 (45%), Positives = 250/397 (62%), Gaps = 14/397 (3%) Frame = +1 Query: 1 KWFGNTRVISEKDLMKFQEY--QNTYDPFQVTLRKTVMPLSLNLHKTKKNKPNFKTLQPY 174 +WFGNTRVI++ L F+E + D +QV LR+ +P+SL K + + Y Sbjct: 78 RWFGNTRVIAQDALNHFREALGETKRDSYQVLLRQNKLPMSLLEENNKIPQVKVVETESY 137 Query: 175 ---FKRKITVKESNIKARTFEELSAQTKTALEEFNAKQ---SMLSIDDDKDEATHGVFRA 336 F K K+ + F EL++ + ++F AK+ + +D EA +F Sbjct: 138 ANTFGPKAQRKKPQLAVGDFAELASAADESQQDFEAKKEEDNSWKVDGWSQEAKEAIFHK 197 Query: 337 GTSNRIWQEVYKVIDSSDVLLYVLDARNPMGMRSPKIELDIRNK--QKKIIFVLNKVDLV 510 G S RIW E+YKVIDSSDV+++VLDAR+P+G R +E I+ + K +IFVLNK DLV Sbjct: 198 GQSKRIWNELYKVIDSSDVVIHVLDARDPLGTRCTSVEQYIKKEAPHKHLIFVLNKCDLV 257 Query: 511 PSGVTRHWEKLLNKEYPTIMFYADVKHPLGKDRLICLLRQFRDKMKDRRQISVGIIGFPN 690 P+ V W K L+++YPT+ F+A + + GK LI LLRQ+ DR+QISVG IG+PN Sbjct: 258 PTWVAAAWVKHLSQDYPTLAFHASITNSFGKGSLIQLLRQYSALHPDRQQISVGFIGYPN 317 Query: 691 TGKSSLINTLKGEKVCKVAPIAGETKVWQYISLMKSIDLIDCPGTV-TSEKDEKLSETDL 867 TGKSS+INTL+ +KVCK API GETKVWQYI+LMK I LIDCPG V S+KD SETD+ Sbjct: 318 TGKSSIINTLRKKKVCKTAPIPGETKVWQYITLMKRIFLIDCPGIVPPSQKD---SETDI 374 Query: 868 ILRNSVRVEKVEDADRHIPAVLNIVNRDVINKLYKINQPWNECEQFLEMVAKSRGRLLKG 1047 + R VRVE V +++IPA+L + + Y+++ W+ +FLE +A+ GRLLKG Sbjct: 375 LFRGVVRVEHVSYPEQYIPALLERCETKHLERTYEVS-GWSNATEFLEKIARKHGRLLKG 433 Query: 1048 GQPDIMTVAKEILKDFNTGKLVWYVKP---ENYDNSK 1149 G+PD +AK IL DFN GK+ W+V P E+ +N K Sbjct: 434 GEPDESGIAKLILNDFNRGKIPWFVPPPQAEDMENVK 470
>sp|Q9C3Z4|NOG2_PNECA Nucleolar GTP-binding protein 2 (Binding-inducible GTPase) Length = 483 Score = 318 bits (814), Expect = 4e-86 Identities = 166/399 (41%), Positives = 255/399 (63%), Gaps = 8/399 (2%) Frame = +1 Query: 1 KWFGNTRVISEKDLMKFQEY--QNTYDPFQVTLRKTVMPLSLNLHKTKKNKPNFKTLQPY 174 +WF NTRVIS+ L F+E + DP +V L++ +P+SL + TK K N ++P+ Sbjct: 65 RWFSNTRVISQDVLNMFRESFAEKLNDPCKVLLKQNKLPMSLLMEPTKTRKANIIDIEPF 124 Query: 175 ---FKRKITVKESNIKARTFEELSAQTKTALEEFNAKQSML-SIDDDKDEATHGVFRAGT 342 F +K K + + A + E LS + E + K S ++D + ++ +F GT Sbjct: 125 DDTFGKKSXRKRAKLYASSIENLSNFAFESYENYIKKNSEYENVDKNIQKSFEAIFSKGT 184 Query: 343 SNRIWQEVYKVIDSSDVLLYVLDARNPMGMRSPKIELDIRNKQ--KKIIFVLNKVDLVPS 516 S RIW E+YK IDSSDV++ +LDARNP+G R +E ++ ++ K +I +LNK DL+P+ Sbjct: 185 SKRIWNELYKXIDSSDVIIQLLDARNPLGTRCKHVEEYLKKEKPHKHMILLLNKCDLIPT 244 Query: 517 GVTRHWEKLLNKEYPTIMFYADVKHPLGKDRLICLLRQFRDKMKDRRQISVGIIGFPNTG 696 TR W K L+KEYPT+ F+A + +P GK LI LLRQF +RRQISVG IG+PNTG Sbjct: 245 WCTREWIKQLSKEYPTLAFHASINNPFGKGSLIQLLRQFSKLHSNRRQISVGFIGYPNTG 304 Query: 697 KSSLINTLKGEKVCKVAPIAGETKVWQYISLMKSIDLIDCPGTVTSEKDEKLSETDLILR 876 KSS+INTL+ +KVC API GETKVWQY+ + I +IDCPG V ++ SET++I++ Sbjct: 305 KSSVINTLRSKKVCNTAPIPGETKVWQYVRMTSKIFMIDCPGIVPPNSND--SETEIIIK 362 Query: 877 NSVRVEKVEDADRHIPAVLNIVNRDVINKLYKINQPWNECEQFLEMVAKSRGRLLKGGQP 1056 ++R+EKV + +++I A+LN+ + + Y+I+ N+ +F+E++A+ G+LLKGG+ Sbjct: 363 GALRIEKVSNPEQYIHAILNLCETKHLERTYQISGWENDSTKFIELLARKTGKLLKGGEV 422 Query: 1057 DIMTVAKEILKDFNTGKLVWYVKPENYDNSKETLKDFDN 1173 D ++AK ++ DF GK+ W++ P +N +K +N Sbjct: 423 DESSIAKMVINDFIRGKIPWFIAPAQ-ENDPSNIKLSNN 460
>sp|Q6FWS1|NOG2_CANGA Nucleolar GTP-binding protein 2 Length = 494 Score = 315 bits (808), Expect = 2e-85 Identities = 182/419 (43%), Positives = 251/419 (59%), Gaps = 21/419 (5%) Frame = +1 Query: 1 KWFGNTRVISEKDLMKFQEY--QNTYDPFQVTLRKTVMPLSLNLHKTKKNKPNFKTLQPY 174 +WFGNTRVIS+ L F++ + D +QV LR+ +P+SL K P + L+ Sbjct: 76 RWFGNTRVISQDALQHFRDALGETQKDTYQVLLRRNKLPMSLLDEKDSTESPRARILETE 135 Query: 175 -----FKRKITVKESNIKARTFEELSAQTKTA------LEEFNAKQSMLSIDDDKDEA-- 315 F K K+ + A + EEL T+ EE A ++ +D++ Sbjct: 136 SFDQTFGPKAQRKKPRVAASSLEELVQATEEENKTYEEKEELKATLGLMGKQEDEENGWT 195 Query: 316 ---THGVFRAGTSNRIWQEVYKVIDSSDVLLYVLDARNPMGMRSPKIELDIRNK--QKKI 480 +F G S RIW E+YKVIDSSDV+++VLDAR+P+G R +E ++ + K + Sbjct: 196 QVTKEAIFSKGQSKRIWNELYKVIDSSDVVIHVLDARDPLGTRCKSVEEYMKKETPHKHL 255 Query: 481 IFVLNKVDLVPSGVTRHWEKLLNKEYPTIMFYADVKHPLGKDRLICLLRQFRDKMKDRRQ 660 I+VLNK DLVP+ V W K L+K+ PT+ F+A + + GK LI LLRQF DR+Q Sbjct: 256 IYVLNKCDLVPTWVAAAWVKHLSKDRPTLAFHASITNSFGKGSLIQLLRQFSQLHTDRKQ 315 Query: 661 ISVGIIGFPNTGKSSLINTLKGEKVCKVAPIAGETKVWQYISLMKSIDLIDCPGTV-TSE 837 ISVG IG+PNTGKSS+INTL+ +KVC+VAPI GETKVWQYI+LMK I LIDCPG V S Sbjct: 316 ISVGFIGYPNTGKSSIINTLRKKKVCQVAPIPGETKVWQYITLMKRIFLIDCPGIVPPST 375 Query: 838 KDEKLSETDLILRNSVRVEKVEDADRHIPAVLNIVNRDVINKLYKINQPWNECEQFLEMV 1017 KD SE D++ R VRVE V +++IP VL + + Y+I+ W + F+EM+ Sbjct: 376 KD---SEEDILFRGVVRVEHVSHPEQYIPGVLKRCQTKHLERTYEIS-GWKDATDFIEML 431 Query: 1018 AKSRGRLLKGGQPDIMTVAKEILKDFNTGKLVWYVKPENYDNSKETLKDFDNKNWLKKQ 1194 A+ +GRLLKGG+PD V+K+IL DFN GK+ W+V P + K+ D + K Q Sbjct: 432 ARKQGRLLKGGEPDESGVSKQILNDFNRGKIPWFVIPPEKEEDKKRKLDEEEVKSAKMQ 490
>sp|P53742|NOG2_YEAST Nucleolar GTP-binding protein 2 Length = 486 Score = 314 bits (804), Expect = 5e-85 Identities = 179/408 (43%), Positives = 247/408 (60%), Gaps = 21/408 (5%) Frame = +1 Query: 1 KWFGNTRVISEKDLMKFQEY--QNTYDPFQVTLRKTVMPLSLNLHKTKKNKPNFKTLQPY 174 +WFGNTRVIS+ L F+ + D +QV LR+ +P+SL K P + L Sbjct: 76 RWFGNTRVISQDALQHFRSALGETQKDTYQVLLRRNKLPMSLLEEKDADESPKARILDTE 135 Query: 175 -----FKRKITVKESNIKARTFEELSAQTKTALEEFNAKQ------SMLSIDDDKDE--- 312 F K K + A E+L T + ++ KQ ++ +DK+ Sbjct: 136 SYADAFGPKAQRKRPRLAASNLEDLVKATNEDITKYEEKQVLDATLGLMGNQEDKENGWT 195 Query: 313 --ATHGVFRAGTSNRIWQEVYKVIDSSDVLLYVLDARNPMGMRSPKIELDIRNK--QKKI 480 A +F G S RIW E+YKVIDSSDV+++VLDAR+P+G R +E ++ + K + Sbjct: 196 SAAKEAIFSKGQSKRIWNELYKVIDSSDVVIHVLDARDPLGTRCKSVEEYMKKETPHKHL 255 Query: 481 IFVLNKVDLVPSGVTRHWEKLLNKEYPTIMFYADVKHPLGKDRLICLLRQFRDKMKDRRQ 660 I+VLNK DLVP+ V W K L+KE PT+ F+A + + GK LI LLRQF DR+Q Sbjct: 256 IYVLNKCDLVPTWVAAAWVKHLSKERPTLAFHASITNSFGKGSLIQLLRQFSQLHTDRKQ 315 Query: 661 ISVGIIGFPNTGKSSLINTLKGEKVCKVAPIAGETKVWQYISLMKSIDLIDCPGTV-TSE 837 ISVG IG+PNTGKSS+INTL+ +KVC+VAPI GETKVWQYI+LMK I LIDCPG V S Sbjct: 316 ISVGFIGYPNTGKSSIINTLRKKKVCQVAPIPGETKVWQYITLMKRIFLIDCPGIVPPSS 375 Query: 838 KDEKLSETDLILRNSVRVEKVEDADRHIPAVLNIVNRDVINKLYKINQPWNECEQFLEMV 1017 KD SE D++ R VRVE V +++IP VL + + Y+I+ W + +F+E++ Sbjct: 376 KD---SEEDILFRGVVRVEHVTHPEQYIPGVLKRCQVKHLERTYEIS-GWKDATEFIEIL 431 Query: 1018 AKSRGRLLKGGQPDIMTVAKEILKDFNTGKLVWYVKPENYDNSKETLK 1161 A+ +GRLLKGG+PD V+K+IL DFN GK+ W+V P + ++ K Sbjct: 432 ARKQGRLLKGGEPDESGVSKQILNDFNRGKIPWFVLPPEKEGEEKPKK 479
>sp|Q7SHR8|NOG2_NEUCR Nucleolar GTP-binding protein 2 Length = 619 Score = 303 bits (777), Expect = 7e-82 Identities = 173/423 (40%), Positives = 254/423 (60%), Gaps = 36/423 (8%) Frame = +1 Query: 1 KWFGNTRVISEKDLMKFQEY--QNTYDPFQVTLRKTVMPLSL---------NLHKTK--- 138 +WF NTRVIS+ L F+E + DP+ V L+ +P+SL H+ K Sbjct: 75 RWFNNTRVISQDTLTSFREAIAEKDKDPYSVLLKSNKLPMSLIRDGPKDALKKHQAKMTI 134 Query: 139 KNKPNFKTLQPYFKRKITVKESNIKARTFEELSAQTKTALEEFNAKQSMLSI-------- 294 +++P +T P +RK + T +L+ ++ +++ + A+ + + Sbjct: 135 ESEPFSQTFGPKAQRK----RPKLSFNTIGDLTEHSEKSMDTYQARLEEIKLLSGASGYG 190 Query: 295 ----DDDKDE-------ATHGVFRAGTSNRIWQEVYKVIDSSDVLLYVLDARNPMGMRSP 441 DDD E A +F G S RIW E+YKVIDSSDV+L+V+DAR+P+G R Sbjct: 191 GGLADDDVQEEDFSVATAKEAIFTKGQSKRIWNELYKVIDSSDVILHVIDARDPLGTRCR 250 Query: 442 KIE--LDIRNKQKKIIFVLNKVDLVPSGVTRHWEKLLNKEYPTIMFYADVKHPLGKDRLI 615 +E L K +IFVLNK+DLVPS W ++L K++PT + +K+P G+ LI Sbjct: 251 HVEKYLATEAPHKHLIFVLNKIDLVPSKTAAAWIRVLQKDHPTCAMRSSIKNPFGRGSLI 310 Query: 616 CLLRQFRDKMKDRRQISVGIIGFPNTGKSSLINTLKGEKVCKVAPIAGETKVWQYISLMK 795 LLRQF KDR+QISVG++G+PN GKSS+IN L+G+ V KVAPI GETKVWQY++LM+ Sbjct: 311 DLLRQFSILHKDRKQISVGLVGYPNVGKSSIINALRGKPVAKVAPIPGETKVWQYVTLMR 370 Query: 796 SIDLIDCPGTVTSEKDEKLSETDLILRNSVRVEKVEDADRHIPAVLNIVNRDVINKLYKI 975 I LIDCPG V +++ + DL+LR VRVE V++ +++IPAVLN V + + Y++ Sbjct: 371 RIYLIDCPGIVPPNQND--TPQDLLLRGVVRVENVDNPEQYIPAVLNKVKPHHMERTYEL 428 Query: 976 NQPWNECEQFLEMVAKSRGRLLKGGQPDIMTVAKEILKDFNTGKLVWYV-KPENYDNSKE 1152 + W + FLEM+A+ GRLLKGG+PD+ VAK +L DF GK+ W+ PE + + Sbjct: 429 -KGWKDHIHFLEMLARKGGRLLKGGEPDVDGVAKMVLNDFMRGKIPWFTPAPEKEEGETD 487 Query: 1153 TLK 1161 T++ Sbjct: 488 TME 490
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 178,577,948
Number of Sequences: 369166
Number of extensions: 3680444
Number of successful extensions: 12021
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 10946
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 11867
length of database: 68,354,980
effective HSP length: 116
effective length of database: 46,925,720
effective search space used: 19943431000
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail