DrC_00273
- Cluster detail
- GO Category : Not Available
- Consensus Sequence :
BLASTX 2.2.13 [Nov-27-2005]
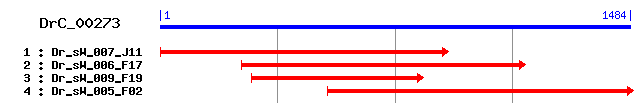
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schäffer, Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997), "Gapped BLAST and PSI-BLAST: a new generation of protein database search programs", Nucleic Acids Res. 25:3389-3402. Query= DrC_00273 (1484 letters) Database: Non-redundant SwissProt sequences 184,735 sequences; 68,354,980 total letters Score E Sequences producing significant alignments: (bits) Value sp|Q16222|UAP1_HUMAN UDP-N-acetylhexosamine pyrophosphoryla... 349 1e-95 sp|Q91YN5|UAP1_MOUSE UDP-N-acetylhexosamine pyrophosphoryla... 348 2e-95 sp|O64765|UAP1_ARATH Probable UDP-N-acetylglucosamine pyrop... 311 3e-84 sp|O74933|UAP1_CANAL UDP-N-acetylglucosamine pyrophosphorylase 300 7e-81 sp|P43123|UAP1_YEAST UDP-N-acetylglucosamine pyrophosphorylase 299 2e-80 sp|O94617|UAP1_SCHPO Probable UDP-N-acetylglucosamine pyrop... 248 3e-65 sp|Q18493|UAP1_CAEEL Probable UDP-N-acetylglucosamine pyrop... 224 5e-58 sp|O59819|UGPA2_SCHPO Probable UTP--glucose-1-phosphate uri... 57 1e-07 sp|P78811|UGPA1_SCHPO Probable UTP--glucose-1-phosphate uri... 54 9e-07 sp|O35156|UGPA1_CRIGR UTP--glucose-1-phosphate uridylyltran... 51 8e-06
>sp|Q16222|UAP1_HUMAN UDP-N-acetylhexosamine pyrophosphorylase (Antigen X) (AGX) (Sperm-associated antigen 2) [Includes: UDP-N-acetylgalactosamine pyrophosphorylase (AGX-1); UDP-N-acetylglucosamine pyrophosphorylase (AGX-2)] Length = 522 Score = 349 bits (896), Expect = 1e-95 Identities = 189/362 (52%), Positives = 244/362 (67%), Gaps = 1/362 (0%) Frame = +2 Query: 26 IPCTVCGSWSDSTPEQLGKYNDIGLKYISNGEYCSILMAGGAGTRLGVKYPKGMYNVGLP 205 +P V GS + +QL + GL IS + +L+AGG GTRLGV YPKGMY+VGLP Sbjct: 72 VpreVLGS-ATRDQDQLQAWESEGLFQISQNKVAVLLLAGGQGTRLGVAYPKGMYDVGLP 130 Query: 206 SGKSLFQLQAERLLRLQHLALMKYGKTAPISWYIMTSDCTADATMEYFKDNNYFGLSPDQ 385 S K+LFQ+QAER+L+LQ +A YG I WYIMTS T ++T E+F + YFGL + Sbjct: 131 SRKTLFQIQAERILKLQQVAEKYYGNKCIIPWYIMTSGRTMESTKEFFTKHKYFGLKKEN 190 Query: 386 IIFFEQFTLPVVGCNGKIILETKCKISKSPDGNGGLYRALYNYKLVDDMKSRGIKYALVY 565 +IFF+Q LP + +GKIILE K K+S +PDGNGGLYRAL +V+DM+ RGI VY Sbjct: 191 VIFFQQGMLPAMSFDGKIILEEKNKVSMAPDGNGGLYRALAAQNIVEDMEQRGIWSIHVY 250 Query: 566 GVDNILVKIVDPVFLGFCVEKGAECAAKVVEKVHPKEAVGIVGKVNGCYQVIEYSEISLE 745 VDNILVK+ DP F+GFC++KGA+C AKVVEK +P E VG+V +V+G YQV+EYSEISL Sbjct: 251 CVDNILVKVADPRFIGFCIQKGADCGAKVVEKTNPTEPVGVVCRVDGVYQVVEYSEISLA 310 Query: 746 TATMRDIKNDKLLYRCGSICTHLFSVDFLSRFCQNNVNNLLLQHIAIKKVPFINIQTGIL 925 TA R + +LL+ G+I H F+V FL R N L H+A KK+P+++ Q G L Sbjct: 311 TAQKRS-SDGRLLFNAGNIANHFFTVPFL-RDVVNVYEPQLQHHVAQKKIPYVDTQ-GQL 367 Query: 926 EIPEKENAIKMEKFVFDVFPLAKTFAIWEVERNREFSALKNAPGSQ-SDCPETAKNDLLA 1102 P+K N IKMEKFVFD+F AK F ++EV R EFS LKNA D P TA++ L++ Sbjct: 368 IKPDKPNGIKMEKFVFDIFQFAKKFVVYEVLREDEFSPLKNADSQNGKDNPTTARHALMS 427 Query: 1103 YH 1108 H Sbjct: 428 LH 429
>sp|Q91YN5|UAP1_MOUSE UDP-N-acetylhexosamine pyrophosphorylase [Includes: UDP-N-acetylgalactosamine pyrophosphorylase ; UDP-N-acetylglucosamine pyrophosphorylase ] Length = 522 Score = 348 bits (893), Expect = 2e-95 Identities = 186/362 (51%), Positives = 243/362 (67%), Gaps = 1/362 (0%) Frame = +2 Query: 26 IPCTVCGSWSDSTPEQLGKYNDIGLKYISNGEYCSILMAGGAGTRLGVKYPKGMYNVGLP 205 +P V GS + EQL + GL IS + +L+AGG GTRLGV YPKGMY+VGLP Sbjct: 72 VPRQVLGS-ATRDQEQLQAWESEGLSQISQNKVAVLLLAGGQGTRLGVSYPKGMYDVGLP 130 Query: 206 SGKSLFQLQAERLLRLQHLALMKYGKTAPISWYIMTSDCTADATMEYFKDNNYFGLSPDQ 385 S K+LFQ+QAER+L+LQ LA +G I WYIMTS T ++T E+F + +FGL + Sbjct: 131 SHKTLFQIQAERILKLQQLAEKHHGNKCTIPWYIMTSGRTMESTKEFFTKHKFFGLKKEN 190 Query: 386 IIFFEQFTLPVVGCNGKIILETKCKISKSPDGNGGLYRALYNYKLVDDMKSRGIKYALVY 565 ++FF+Q LP + +GKIILE K K+S +PDGNGGLYRAL +V+DM+ RGI VY Sbjct: 191 VVFFQQGMLPAMSFDGKIILEEKNKVSMAPDGNGGLYRALAAQNIVEDMEQRGICSIHVY 250 Query: 566 GVDNILVKIVDPVFLGFCVEKGAECAAKVVEKVHPKEAVGIVGKVNGCYQVIEYSEISLE 745 VDNILVK+ DP F+GFC++KGA+C AKVVEK +P E VG+V +V+G YQV+EYSEISL Sbjct: 251 CVDNILVKVADPRFIGFCIQKGADCGAKVVEKTNPTEPVGVVCRVDGVYQVVEYSEISLA 310 Query: 746 TATMRDIKNDKLLYRCGSICTHLFSVDFLSRFCQNNVNNLLLQHIAIKKVPFINIQTGIL 925 TA R + +LL+ G+I H F+V FL + N L H+A KK+P+++ Q G Sbjct: 311 TAQRRS-SDGRLLFNAGNIANHFFTVPFL-KDVVNVYEPQLQHHVAQKKIPYVDSQ-GYF 367 Query: 926 EIPEKENAIKMEKFVFDVFPLAKTFAIWEVERNREFSALKNAPGSQ-SDCPETAKNDLLA 1102 P+K N IKMEKFVFD+F AK F ++EV R EFS LKNA D P TA++ L++ Sbjct: 368 IKPDKPNGIKMEKFVFDIFQFAKKFVVYEVLREDEFSPLKNADSQNGKDNPTTARHALMS 427 Query: 1103 YH 1108 H Sbjct: 428 LH 429
>sp|O64765|UAP1_ARATH Probable UDP-N-acetylglucosamine pyrophosphorylase Length = 502 Score = 311 bits (797), Expect = 3e-84 Identities = 171/405 (42%), Positives = 249/405 (61%), Gaps = 8/405 (1%) Frame = +2 Query: 26 IPCTVCGSWSDSTPEQLGKYNDIGLKYISNGEYCSILMAGGAGTRLGVKYPKGMYNVGLP 205 +P + + T E K+ +GLK I G+ +L++GG GTRLG PKG YN+GLP Sbjct: 93 VPENCVSTVEERTKEDREKWWKMGLKAIYEGKLGVVLLSGGQGTRLGSSDPKGCYNIGLP 152 Query: 206 SGKSLFQLQAERLLRLQHLALMKYGKTAP-----ISWYIMTSDCTADATMEYFKDNNYFG 370 SGKSLFQ+QAER+L +Q LA + +P I WYIMTS T + T ++FK + YFG Sbjct: 153 SGKSLFQIQAERILCVQRLASQAMSEASPTRPVTIQWYIMTSPFTHEPTQKFFKSHKYFG 212 Query: 371 LSPDQIIFFEQFTLPVVGCNGKIILETKCKISKSPDGNGGLYRALYNYKLVDDMKSRGIK 550 L PDQ+ FF+Q TLP + +GK I+ET +SK+PDGNGG+Y AL + +L++DM SRGIK Sbjct: 213 LEPDQVTFFQQGTLPCISKDGKFIMETPFSLSKAPDGNGGVYTALKSSRLLEDMASRGIK 272 Query: 551 YALVYGVDNILVKIVDPVFLGFCVEKGAECAAKVVEKVHPKEAVGIV---GKVNGCYQVI 721 Y YGVDN+LV++ DP FLG+ ++K A AAKVV K +P+E VG+ GK G V+ Sbjct: 273 YVDCYGVDNVLVRVADPTFLGYFIDKSAASAAKVVRKAYPQEKVGVFVRRGK-GGPLTVV 331 Query: 722 EYSEISLETATMRDIKNDKLLYRCGSICTHLFSVDFLSRFCQNNVNNLLLQHIAIKKVPF 901 EY+E+ A+ + + +L Y ++C H+F++DFL++ N + + H+A KK+P Sbjct: 332 EYTELDQSMASATNQQTGRLQYCWSNVCLHMFTLDFLNQVA-NGLEKDSVYHLAEKKIPS 390 Query: 902 INIQTGILEIPEKENAIKMEKFVFDVFPLAKTFAIWEVERNREFSALKNAPGSQSDCPET 1081 IN +K+E+F+FD FP A + A++EV R EF+ +KNA GS D PE+ Sbjct: 391 INGDI---------VGLKLEQFIFDCFPYAPSTALFEVLREEEFAPVKNANGSNYDTPES 441 Query: 1082 AKNDLLAYHRQXXXXXXXXXXXXKEKVKKTIIELSPLLTYSGEDM 1216 A+ +L H + + T +E+SPL +Y+GE++ Sbjct: 442 ARLLVLRLHTR-WVIAAGGFLTHSVPLYATGVEVSPLCSYAGENL 485
>sp|O74933|UAP1_CANAL UDP-N-acetylglucosamine pyrophosphorylase Length = 486 Score = 300 bits (768), Expect = 7e-81 Identities = 170/370 (45%), Positives = 237/370 (64%), Gaps = 10/370 (2%) Frame = +2 Query: 20 TTIPCTVCGSWSDSTPEQLGKYNDIGLKYISNGEYCSILMAGGAGTRLGVKYPKGMYNVG 199 T +P S D + + L + ++GLK I NGE +LMAGG GTRLG PKG +N+ Sbjct: 70 TQLPNEQTASTLDLSKDILQNWTELGLKAIGNGEVAVLLMAGGQGTRLGSSAPKGCFNIE 129 Query: 200 LPSGKSLFQLQAERLLRLQHLA--LMKYGKTAPISWYIMTSDCTADATMEYFKDNNYFGL 373 LPS KSLFQ+QAE++L+++ LA +K K I+WYIMTS T +AT +F +NNYFGL Sbjct: 130 LPSQKSLFQIQAEKILKIEQLAQQYLKSTKKPIINWYIMTSGPTRNATESFFIENNYFGL 189 Query: 374 SPDQIIFFEQFTLPVVGCNG-KIILETKCKISKSPDGNGGLYRALYNYKLVDDMKSRGIK 550 + Q+IFF Q TLP G KI+LE+K I +SPDGNGGLY+AL + ++DD+ S+GIK Sbjct: 190 NSHQVIFFNQGTLPCFNLQGNKILLESKNSICQSPDGNGGLYKALKDNGILDDLNSKGIK 249 Query: 551 YALVYGVDNILVKIVDPVFLGFCVEKGAECAAKVVEKVHPKEAVGIV--GKVNGCYQVIE 724 + +Y VDN LVK+ DP+F+GF + K + A KVV K E+VG++ + N VIE Sbjct: 250 HIHMYCVDNCLVKVADPIFIGFAIAKKFDLATKVVRKRDANESVGLIVLDQDNQKPCVIE 309 Query: 725 YSEISLETATMRDIK-NDKLLYRCGSICTHLFSVDFLSRFCQNNVNN--LLLQHIAIKKV 895 YSEIS E A +D + + KL R +I H +SV+FL++ +++ L HIA KK+ Sbjct: 310 YSEISQELANKKDPQDSSKLFLRAANIVNHYYSVEFLNKMIPKWISSQKYLPFHIAKKKI 369 Query: 896 PFINIQTGILEIPEKENAIKMEKFVFDVFPLAK--TFAIWEVERNREFSALKNAPGSQSD 1069 P +N++ G P + N IK+E+F+FDVFP + F EV+R EFS LKNA G+++D Sbjct: 370 PSLNLENGEFYKPTEPNGIKLEQFIFDVFPSVELNKFGCLEVDRLDEFSPLKNADGAKND 429 Query: 1070 CPETAKNDLL 1099 P T +N L Sbjct: 430 TPTTCRNHYL 439
>sp|P43123|UAP1_YEAST UDP-N-acetylglucosamine pyrophosphorylase Length = 477 Score = 299 bits (765), Expect = 2e-80 Identities = 187/407 (45%), Positives = 244/407 (59%), Gaps = 8/407 (1%) Frame = +2 Query: 80 KYNDIGLKYISNGEYCSILMAGGAGTRLGVKYPKGMYNVGLPSGKSLFQLQAERLLRLQH 259 +Y +GL+ I GE ILMAGG GTRLG PKG Y++GLPS KSLFQ+QAE+L+RLQ Sbjct: 90 EYWRLGLEAIGKGEVAVILMAGGQGTRLGSSQPKGCYDIGLPSKKSLFQIQAEKLIRLQD 149 Query: 260 LALMKYGKTAPISWYIMTSDCTADATMEYFKDNNYFGLSPDQIIFFEQFTLPVVGCNGK- 436 M K I WYIMTS T AT YF+++NYFGL+ +QI FF Q TLP GK Sbjct: 150 ---MVKDKKVEIPWYIMTSGPTRAATEAYFQEHNYFGLNKEQITFFNQGTLPAFDLTGKH 206 Query: 437 IILETKCKISKSPDGNGGLYRALYNYKLVDDMKSRGIKYALVYGVDNILVKIVDPVFLGF 616 +++ +S+SPDGNGGLYRA+ KL +D RGIK+ +Y VDN+L KI DPVF+GF Sbjct: 207 FLMKDPVNLSQSPDGNGGLYRAIKENKLNEDFDRRGIKHVYMYCVDNVLSKIADPVFIGF 266 Query: 617 CVEKGAECAAKVVEKVHPKEAVGIVGKVNGCYQVIEYSEISLETATMRDIKNDKLLYRCG 796 ++ G E A K V K E+VG++ N VIEYSEIS E A +D K+ L R G Sbjct: 267 AIKHGFELATKAVRKRDAHESVGLIATKNEKPCVIEYSEISNELAEAKD-KDGLLKLRAG 325 Query: 797 SICTHLFSVDFLSR----FCQNNVNNLLLQHIAIKKVPFINIQTGILEIPEKENAIKMEK 964 +I H + VD L R +C+N + HIA KK+P + TG P + N IK+E+ Sbjct: 326 NIVNHYYLVDLLKRDLDQWCEN-----MPYHIAKKKIPAYDSVTGKYTKPTEPNGIKLEQ 380 Query: 965 FVFDVF---PLAKTFAIWEVERNREFSALKNAPGSQSDCPETAKNDLLAYHRQXXXXXXX 1135 F+FDVF PL K F EV+R +EFS LKN PGS++D PET++ LAY + Sbjct: 381 FIFDVFDTVPLNK-FGCLEVDRCKEFSPLKNGPGSKNDNPETSR---LAYLKLGTSWLED 436 Query: 1136 XXXXXKEKVKKTIIELSPLLTYSGEDMGNISVLIKNSGDESIVQLEK 1276 K+ V ++E+S L+Y+GE N+S D S + LEK Sbjct: 437 AGAIVKDGV---LVEVSSKLSYAGE---NLSQFKGKVFDRSGIVLEK 477
>sp|O94617|UAP1_SCHPO Probable UDP-N-acetylglucosamine pyrophosphorylase Length = 475 Score = 248 bits (633), Expect = 3e-65 Identities = 134/343 (39%), Positives = 203/343 (59%), Gaps = 7/343 (2%) Frame = +2 Query: 95 GLKYISNGEYCSILMAGGAGTRLGVKYPKGMYNVGLPSGKSLFQLQAERL---LRLQHLA 265 GL+ I+ G ++++AGG GTRLG PKG + +GLP+ S+F+LQA+++ L L A Sbjct: 89 GLREIARGHVAALVLAGGQGTRLGFAGPKGCFRLGLPNNPSIFELQAQKIKKSLALARAA 148 Query: 266 LMKYGKTAPISWYIMTSDCTADATMEYFKDNNYFGLSPDQIIFFEQFTLPVVGCNGKIIL 445 + I WYIM S+CT++ T+ +FK+N++FG+ + FF+Q LP + +G+++ Sbjct: 149 FPDQEASISIPWYIMVSECTSEETISFFKENDFFGIDKKDVFFFQQGVLPCLDISGRVLF 208 Query: 446 ETKCKISKSPDGNGGLYRALYNYKLVDDMKSRGIKYALVYGVDNILVKIVDPVFLGFCVE 625 E+ ++ +P+GNGG+Y AL + ++DM RGI + Y VDN+LV VDPVF+G Sbjct: 209 ESDSSLAWAPNGNGGIYEALLSSGALNDMNRRGILHITAYSVDNVLVLPVDPVFIGMATT 268 Query: 626 KGAECAAKVVEKVHPKEAVGIVGKVNGCYQVIEYSEISLET--ATMRDIKNDKLLYRCGS 799 K E A K VEK+ P E VG++ + V+EYSEIS E AT + LL R + Sbjct: 269 KKLEVATKTVEKIDPAEKVGLLVSSHNHPCVVEYSEISDEACKATENVDGHKHLLLRAAN 328 Query: 800 ICTHLFSVDFLSRFCQNNVNNLLLQHIAIKKVPFINIQTGILEIPEKENAIKMEKFVFDV 979 I H FS DFL + + ++ L H+A KK+PF ++ + P N K+E F+FD+ Sbjct: 329 IAYHYFSFDFLQKASLH--SSTLPIHLACKKIPFYDVTSHHYTTPLNPNGYKLESFIFDL 386 Query: 980 FP--LAKTFAIWEVERNREFSALKNAPGSQSDCPETAKNDLLA 1102 FP + F ++V R FS LKN+ S +D ET ND+L+ Sbjct: 387 FPSVSVENFGCFQVPRRTSFSPLKNSSKSPNDNHETCVNDILS 429
>sp|Q18493|UAP1_CAEEL Probable UDP-N-acetylglucosamine pyrophosphorylase Length = 484 Score = 224 bits (571), Expect = 5e-58 Identities = 146/379 (38%), Positives = 210/379 (55%), Gaps = 5/379 (1%) Frame = +2 Query: 95 GLKYISNGEYCSILMAGGAGTRLGVKYPKGMYNVGLPS--GKSLFQLQAERLLRLQHLA- 265 G+ I GE C+I++AGG TRLG PKG +G+ + G SL +QA ++ LQ LA Sbjct: 93 GMDAIGRGEVCAIVLAGGQATRLGSSQPKGTIPLGINASFGDSLLGIQAAKIALLQALAG 152 Query: 266 LMKYGKTAPISWYIMTSDCTADATMEYFKD-NNYFGLSPD-QIIFFEQFTLPVVGCNGKI 439 ++ I W +MTS T +AT E+ K + G D QI F Q + G Sbjct: 153 EREHQNPGKIHWAVMTSPGTEEATREHVKKLAAHHGFDFDEQITIFSQDEIAAYDEQGNF 212 Query: 440 ILETKCKISKSPDGNGGLYRALYNYKLVDDMKSRGIKYALVYGVDNILVKIVDPVFLGFC 619 +L TK + +P+GNGGLY A+ + + ++++GIKY VY VDNIL K+ DP F+GF Sbjct: 213 LLGTKGSVVAAPNGNGGLYSAISAH--LPRLRAKGIKYFHVYCVDNILCKVADPHFIGFA 270 Query: 620 VEKGAECAAKVVEKVHPKEAVGIVGKVNGCYQVIEYSEISLETATMRDIKNDKLLYRCGS 799 + A+ A K V K E VG V G +V+EYSE+ E A + + K L+ GS Sbjct: 271 ISNEADVATKCVPK-QKGELVGSVCLDRGLPRVVEYSELGAELAEQKT-PDGKYLFGAGS 328 Query: 800 ICTHLFSVDFLSRFCQNNVNNLLLQHIAIKKVPFINIQTGILEIPEKENAIKMEKFVFDV 979 I H F++DF+ R C + ++ L H A KK+ ++N Q I++ PEK N IK+E+F+FDV Sbjct: 329 IANHFFTMDFMDRVC--SPSSRLPYHRAHKKISYVNEQGTIVK-PEKPNGIKLEQFIFDV 385 Query: 980 FPLAKTFAIWEVERNREFSALKNAPGSQSDCPETAKNDLLAYHRQXXXXXXXXXXXXKEK 1159 F L+K F IWEV RN EFS LKNA +DC T + DL ++ K Sbjct: 386 FELSKRFFIWEVARNEEFSPLKNAQSVGTDCLSTCQRDLSNVNK-----LWLERVQAKVT 440 Query: 1160 VKKTIIELSPLLTYSGEDM 1216 + I L +++Y+GE++ Sbjct: 441 ATEKPIYLKTIVSYNGENL 459
>sp|O59819|UGPA2_SCHPO Probable UTP--glucose-1-phosphate uridylyltransferase (UDP-glucose pyrophosphorylase) (UDPGP) (UGPase) Length = 499 Score = 57.0 bits (136), Expect = 1e-07 Identities = 58/238 (24%), Positives = 105/238 (44%), Gaps = 14/238 (5%) Frame = +2 Query: 65 PEQLGKYNDIGL-----KYISNGEYCSILMAGGAGTRLGVKYPKGMYNVGLPSGKSLFQL 229 PE + Y D+ L KY++ + + GG G LGV YPK M V +S L Sbjct: 81 PEDMIDYGDLPLCKNAGKYLNR--LAVVKLNGGMGNALGVNYPKAMIEV--RDNQSFLDL 136 Query: 230 QAERLLRLQHLALMKYGKTAPISWYIMTSDCTADATMEYFKDNNYFGLSPDQIIFFEQFT 409 ++ L +Y + P + +M S T D T + + Y G D I FEQ Sbjct: 137 SIRQIEYLNR----RYDVSVP--FILMNSYDTNDETCKVLR--KYAGCKID-ISTFEQSR 187 Query: 410 LPVVGCNGKIILETKCKISKS---------PDGNGGLYRALYNYKLVDDMKSRGIKYALV 562 P ++ ++++ + K+ P G+G ++ AL + ++ + ++G Y V Sbjct: 188 YP------RVFVDSQLPVPKAAPSPIEEWYPPGHGDIFDALVHSGTIERLLAQGKDYLFV 241 Query: 563 YGVDNILVKIVDPVFLGFCVEKGAECAAKVVEKVHPKEAVGIVGKVNGCYQVIEYSEI 736 +DN L VD L ++ E + ++ +K VGI+ +G +++E +++ Sbjct: 242 SNIDN-LGASVDLNILSHVIDNQIEYSMEITDKTKADIKVGILVNQDGLLRLLETNQV 298
>sp|P78811|UGPA1_SCHPO Probable UTP--glucose-1-phosphate uridylyltransferase (UDP-glucose pyrophosphorylase) (UDPGP) (UGPase) Length = 506 Score = 54.3 bits (129), Expect = 9e-07 Identities = 71/306 (23%), Positives = 122/306 (39%), Gaps = 9/306 (2%) Frame = +2 Query: 65 PEQLGKYNDI----GLKYISNGEYCSILMAGGAGTRLGVKYPKGMYNVGLPSGKSLFQLQ 232 PEQ+ +Y+ I GL + + + GG GT +G PK + V G S L Sbjct: 87 PEQVVEYDTITEAGGLSRDYLNKLAVLKLNGGLGTTMGCVGPKSIIEVR--DGNSFLDLS 144 Query: 233 AERLLRLQHLALMKYGKTAPISWYIMTSDCTADATMEYFKDNNYFGLSPDQIIFFEQFTL 412 ++ L KY P + +M S T +AT + K + I+ F Q Sbjct: 145 VRQIEHLNR----KYNVNVP--FVLMNSFNTDEATAKVIKKYEAHKID---ILTFNQSRY 195 Query: 413 PVVGCNGKIILETKCKISKS-----PDGNGGLYRALYNYKLVDDMKSRGIKYALVYGVDN 577 P V + + +L + P G+G ++ AL N ++D + ++G +Y V +DN Sbjct: 196 PRV--HKETLLPVPHTADSAIDEWYPPGHGDVFEALTNSGIIDTLIAQGKEYLFVSNIDN 253 Query: 578 ILVKIVDPVFLGFCVEKGAECAAKVVEKVHPKEAVGIVGKVNGCYQVIEYSEISLETATM 757 L +VD L VE AE ++ K G + +G +++E +++ + Sbjct: 254 -LGAVVDLNILNHMVETNAEYLMELTNKTKADVKGGTLIDYDGNVRLLEIAQVPPQ---- 308 Query: 758 RDIKNDKLLYRCGSICTHLFSVDFLSRFCQNNVNNLLLQHIAIKKVPFINIQTGILEIPE 937 H+ + +F N NNL ++K+V +N +EI Sbjct: 309 -----------------HVEEFKSIKKFKYFNTNNLWFHLPSVKRV--VNNHELSMEIIP 349 Query: 938 KENAIK 955 + IK Sbjct: 350 NKKTIK 355
>sp|O35156|UGPA1_CRIGR UTP--glucose-1-phosphate uridylyltransferase 1 (UDP-glucose pyrophosphorylase 1) (UDPGP 1) (UGPase 1) Length = 508 Score = 51.2 bits (121), Expect = 8e-06 Identities = 65/292 (22%), Positives = 114/292 (39%), Gaps = 12/292 (4%) Frame = +2 Query: 131 ILMAGGAGTRLGVKYPKGMYNVGLPSGKSLFQLQAERLLRLQHLALMKYGKTAPISWYIM 310 + + GG GT +G K PK + +G+ + + L +++ L Y P+ +M Sbjct: 111 VKLNGGLGTSMGCKGPKSL--IGVRNENTFLDLTVQQIEHLNK----SYNTDVPL--VLM 162 Query: 311 TSDCTADATMEYFKDNNYFGLSPDQIIFFEQFTLPVVGCNGKIILETKCKISKS------ 472 S T + T + + N+ + +I F Q P + N + +L +S S Sbjct: 163 NSFNTDEDTKKILQKYNHCRV---KIYTFNQSRYPRI--NKESLLPVAKDVSSSGESTEA 217 Query: 473 --PDGNGGLYRALYNYKLVDDMKSRGIKYALVYGVDNILVKIVDPVFLGFCVE----KGA 634 P G+G +Y + YN L+D G +Y V +DN L VD L + K Sbjct: 218 WYPPGHGDIYASFYNSGLLDTFLEEGKEYIFVSNIDN-LGATVDLYILNHLMNPPNGKRC 276 Query: 635 ECAAKVVEKVHPKEAVGIVGKVNGCYQVIEYSEISLETATMRDIKNDKLLYRCGSICTHL 814 E +V K G + + G +++E +++ H+ Sbjct: 277 EFVMEVTNKTRADVKGGTLTQYEGKLRLVEIAQVPK---------------------AHV 315 Query: 815 FSVDFLSRFCQNNVNNLLLQHIAIKKVPFINIQTGILEIPEKENAIKMEKFV 970 +S+F N NNL + A+K++ +++NAI ME V Sbjct: 316 DEFKSVSKFKIFNTNNLWISLAAVKRL-------------QEQNAIDMEIIV 354
Database: Non-redundant SwissProt sequences
Posted date: Dec 6, 2005 7:40 AM
Number of letters in database: 68,354,980
Number of sequences in database: 184,735
Database: swissprot.01
Posted date: Dec 6, 2005 8:18 AM
Number of letters in database: 66,202,850
Number of sequences in database: 184,431
Lambda K H
0.318 0.134 0.401
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 170,539,377
Number of Sequences: 369166
Number of extensions: 3569696
Number of successful extensions: 9423
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 8818
Number of HSP's successfully gapped in prelim test: 0
Number of HSP's that attempted gapping in prelim test: 0
Number of HSP's gapped (non-prelim): 9387
length of database: 68,354,980
effective HSP length: 115
effective length of database: 47,110,455
effective search space used: 17854862445
frameshift window, decay const: 40, 0.1
T: 12
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
Cluster detail